The Adenosine A1 Receptor in Breast Cancer: Emerging Target for Therapeutic Intervention
This article provides a comprehensive preliminary investigation into the role of the Adenosine A1 Receptor (A1R) in breast cancer.
The Adenosine A1 Receptor in Breast Cancer: Emerging Target for Therapeutic Intervention
Abstract
This article provides a comprehensive preliminary investigation into the role of the Adenosine A1 Receptor (A1R) in breast cancer. Targeting the adenosinergic pathway, particularly the A1R, represents a promising new frontier in oncology. We explore the foundational biology of A1R and its expression within the breast tumor microenvironment, review cutting-edge methodological approaches for investigating this target—from bioinformatics to molecular docking and in vitro validation—and address key challenges in drug development, such as achieving subtype selectivity. Finally, we synthesize validation strategies and comparative analyses with other adenosine receptors, offering a consolidated perspective for researchers and drug development professionals aiming to translate A1R modulation into effective breast cancer therapies.
Unraveling the Role of the A1 Adenosine Receptor in Breast Cancer Biology
Adenosine A1 Receptor Signaling and Downstream Pathways in Cancer
The adenosine A1 receptor (A1AR) is a G-protein-coupled receptor (GPCR) that has emerged as a significant player in cancer biology, particularly in breast cancer. Within the tumor microenvironment, extracellular adenosine concentrations rise, often due to hypoxia and the enzymatic breakdown of ATP by CD39 and CD73 [1]. This adenosine can then activate A1AR, initiating signaling cascades that influence critical cellular processes such as proliferation, apoptosis, and migration. The receptor's role is complex, acting as both a target and a regulator of established oncogenic pathways, including those driven by estrogen receptor-alpha (ERα) [2]. This preliminary investigation frames the A1AR within the context of breast cancer research, synthesizing current understanding of its signaling mechanisms, downstream effects, and emerging potential as a therapeutic target. The following sections provide an in-depth technical analysis of the molecular pathways involved, supported by experimental data and protocols relevant to researchers and drug development professionals.
Molecular Biology of the Adenosine A1 Receptor
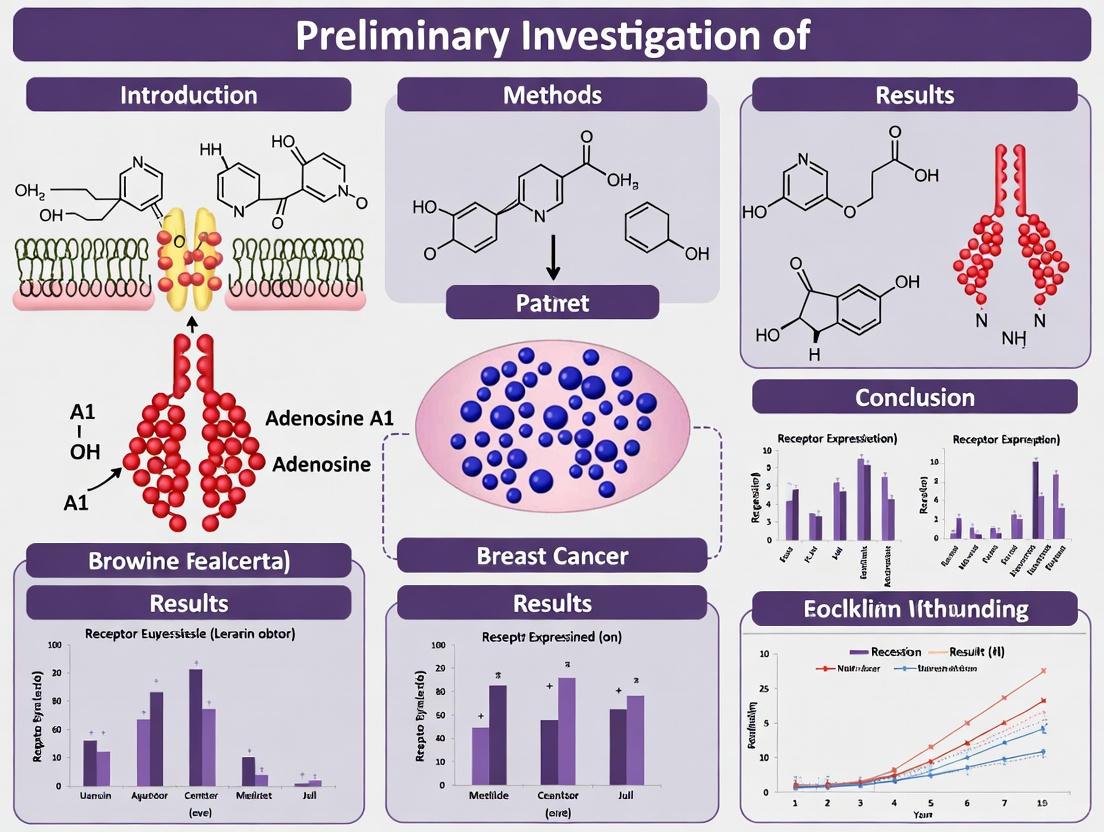
The A1AR is a class A GPCR characterized by its high affinity for adenosine. Its structure consists of seven transmembrane domains, and it primarily couples to the Gi/o family of G proteins [1]. Upon agonist binding, the canonical signaling pathway involves the inhibition of adenylate cyclase (AC), leading to a reduction in intracellular cyclic AMP (cAMP) levels [3] [1]. Beyond this primary pathway, A1AR activation can also stimulate phospholipase C (PLC), leading to the generation of inositol trisphosphate (IP3) and diacylglycerol (DAG), which in turn modulate intracellular calcium levels and protein kinase C (PKC) activity [1]. In the context of cancer, and specifically breast cancer, A1AR is frequently overexpressed, and its expression is functionally linked to cell viability and the suppression of apoptosis [4] [5]. Intriguingly, in ERα-positive breast cancer cells, A1AR expression is upregulated by estradiol (E2), forming a feed-forward loop that promotes cancer growth [2] [6].
Downstream Signaling Pathways and Functional Outcomes
The activation of A1AR initiates a network of downstream signaling events that culminate in distinct functional outcomes in cancer cells. The diagram below illustrates the core signaling pathways and their biological consequences.
The core A1AR signaling network illustrates how receptor activation influences key cellular decisions. The primary pathway involves the inhibition of adenylate cyclase and a subsequent decrease in cAMP, which reduces Protein Kinase A (PKA) activity. This suppression of the cAMP/PKA axis promotes proliferation and suppresses apoptosis [7] [1]. Concurrently, A1AR can stimulate PLC, leading to PKC activation and calcium release, which further drives proliferation and migration [1]. A critical regulatory node is the reciprocal relationship with ERα, where A1AR enhances ERα transcriptional activity, and in turn, is upregulated by E2 [2] [6]. Furthermore, pharmacological inhibition of A1AR leads to the upregulation of the tumor suppressor p53 and executioner caspases, pushing the cell toward apoptosis [4] [8] [5].
Key Functional Consequences in Cancer
Promotion of Cell Proliferation: The A1AR-driven reduction in cAMP and enhancement of ERα activity creates a synergistic pro-proliferative signal. Studies show that A1AR ablation or pharmacological inhibition reduces basal and E2-dependent proliferation in ERα-positive breast cancer cells [2] [6]. Furthermore, overexpression of A1AR in an ERα-negative cell line was sufficient to induce proliferation [2].
Inhibition of Apoptosis: A1AR signaling confers resistance to programmed cell death. Research indicates that the A1AR agonist CPA increases cell viability and reduces apoptosis, an effect correlated with the down-regulation of p53 and caspases 3, 8, and 9 [4] [5]. Conversely, the antagonist DPCPX induces apoptosis by upregulating these key apoptotic molecules [4] [8] [5].
Modulation of the Tumor Microenvironment: While this review focuses on breast cancer cells, it is noted that A1AR on host cells, such as microglial cells in glioblastoma models, can modulate tumor growth [3]. This highlights the potential for A1AR signaling to influence the broader tumor microenvironment.
Quantitative Data on A1AR Modulation in Breast Cancer Models
The effects of A1AR modulation have been quantified in various in vitro studies, primarily using the MCF-7 breast cancer cell line. The tables below consolidate key quantitative findings for easy comparison.
Table 1: Impact of A1AR Ligands on Cell Viability and Apoptosis in MCF-7 Cells
| Ligand | Role | Concentration | Exposure Time | Key Quantitative Outcome | Source |
|---|---|---|---|---|---|
| DPCPX | Antagonist | 87 nM | 72 h | Significantly induced apoptosis; reduced cell viability | [4] [5] |
| CPA | Agonist | 180 μM | 72 h | Increased cell viability; reduced apoptosis | [4] [5] |
| ODC-MPI-2 | Dual A1AR/ODC Inhibitor | Varies | 48-72 h | Inhibited growth, proliferation, and migration/invasion | [7] |
Table 2: Gene Expression Changes Following A1AR Antagonism with DPCPX
| Gene | Function | Expression Change | Experimental Method | Biological Consequence | Source |
|---|---|---|---|---|---|
| TP53 (p53) | Tumor suppressor | Upregulated | Real-time PCR | Promotion of cell cycle arrest and apoptosis | [4] [8] [5] |
| CASP3 | Executioner caspase | Upregulated | Real-time PCR | Induction of apoptotic cell death | [4] [5] |
| CASP8 | Extrinsic pathway caspase | Upregulated | Real-time PCR | Activation of extrinsic apoptosis | [4] [5] |
| CASP9 | Intrinsic pathway caspase | Upregulated | Real-time PCR | Activation of intrinsic apoptosis | [4] [5] |
| ERα | Estrogen receptor | Downregulated (via siRNA) | Western Blot / PCR | Reduced E2/ERα transcriptional activity | [2] [6] |
Detailed Experimental Protocols for Key Assays
To investigate A1AR signaling in cancer, a combination of molecular, cellular, and computational approaches is employed. The workflow below outlines a typical integrated research strategy.
This experimental workflow begins with target identification and proceeds through computational and laboratory validation stages. Below are detailed protocols for key methodologies.
In Silico Target Screening and Molecular Docking
Objective: To identify A1AR as a potential therapeutic target and evaluate compound binding affinity.
- Target Prediction: Input the chemical structures of known active compounds into the SwissTargetPrediction database (http://swisstargetprediction.ch), specifying "Homo sapiens" as the species to generate a list of potential protein targets [9].
- Intersection Analysis: Use a online tool (e.g., Venny) to perform an intersection analysis of targets predicted for multiple active compounds to identify common targets, such as A1AR [9].
- Ligand Preparation: Create a ligand library and optimize 3D structures using software like Discovery Studio. Perform energy minimization and assign partial charges [9].
- Molecular Docking: Dock the prepared ligands into the binding site of the A1AR structure (e.g., PDB ID: 7LD3) using a docking program (e.g., CHARMM within Discovery Studio). Use the LibDock score to evaluate binding poses, typically considering scores >130 as indicative of strong binding affinity [9].
Molecular Dynamics (MD) Simulation
Objective: To assess the stability and dynamics of the A1AR-ligand complex over time.
- System Setup: Use GROMACS 2020.3 with the AMBER99SB-ILDN force field for the protein. Generate ligand parameters using the GAFF force field via ACPYPE. Solvate the system in a cubic box with TIP3P water molecules and add ions (e.g., Cl-) to achieve electrical neutrality [9].
- Energy Minimization: Perform an initial energy minimization step using the steepest descent algorithm to relieve any steric clashes.
- Equilibration: Conduct a 150 ps restrained MD simulation under NVT (constant Number of particles, Volume, and Temperature) and NPT (constant Number of particles, Pressure, and Temperature) ensembles to stabilize the system temperature (298.15 K) and pressure (1 bar) [9].
- Production Run: Execute an unrestricted MD simulation for a defined period (e.g., 15 ns or longer) with a time step of 0.002 ps. Analyze the resulting trajectory (e.g., using VMD 1.9.3) for root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), and specific ligand-receptor interactions across simulation frames [9].
In Vitro Analysis of Apoptosis and Gene Expression
Objective: To quantify the effects of A1AR ligands on apoptosis and relevant gene expression in breast cancer cell lines.
- Cell Culture and Treatment: Culture MCF-7 cells in DMEM/F12 medium supplemented with 10% FBS and antibiotics at 37°C in a 5% CO2 humidified atmosphere. Treat cells at ~80% confluence with specific A1AR ligands (e.g., CPA or DPCPX) at their predetermined IC50 concentrations for 24, 48, and 72 hours [4] [5].
- MTT Viability Assay: Seed 10^4 cells per well in a 24-well plate. After treatment, add MTT solution to each well and incubate for 3 hours. Lyse the formed formazan crystals with DMSO and measure the optical density at a specific wavelength (e.g., 570 nm) using a plate reader. Calculate cell viability as a percentage of the control group [4] [5].
- Annexin V/Propidium Iodide (PI) Apoptosis Assay: Harvest at least 4 × 10^5 treated cells, wash with PBS, and resuspend in binding buffer. Stain cells with Annexin V-FITC and PI according to the manufacturer's protocol. Analyze the stained cells using a flow cytometer (e.g., FACScan) to distinguish between live (Annexin V-/PI-), early apoptotic (Annexin V+/PI-), late apoptotic (Annexin V+/PI+), and necrotic (Annexin V-/PI+) cell populations [4] [5].
- Real-time PCR for Gene Expression: Extract total RNA from treated cells using a kit (e.g., RNeasy Mini Kit). Reverse-transcribe 100 ng of RNA into cDNA. Perform real-time PCR using SYBR Green Master Mix and specific primers for p53, caspases 3, 8, 9, and an endogenous control (e.g., GAPDH). Analyze the data using the comparative CT (ΔΔCT) method to determine relative gene expression levels [4] [5].
The Scientist's Toolkit: Key Research Reagents
Table 3: Essential Reagents for Investigating A1AR in Cancer
| Reagent / Tool | Function / Role | Example Usage in Research |
|---|---|---|
| DPCPX | Selective A1AR antagonist | Used to block A1AR activity, leading to reduced proliferation and induced apoptosis in MCF-7 cells; IC50 ~87 nM [4] [5] [7]. |
| CPA (N6-Cyclopentyladenosine) | Selective A1AR agonist | Used to activate A1AR signaling, resulting in increased cell viability and suppressed apoptosis; IC50 ~180 μM [4] [5]. |
| ODC-MPI-2 | Dual A1AR and ODC inhibitor | Used to simultaneously target polyamine synthesis and A1AR signaling, showing synergistic antitumor effects in MCF-7 cells [7]. |
| DFMO (α-Difluoromethylornithine) | Irreversible ODC inhibitor | Used in combination with DPCPX to demonstrate synergistic growth inhibition of MCF-7 cells [7]. |
| siRNA targeting Adora1 | Gene silencing tool | Used to ablate A1AR expression, confirming its role in basal and E2-dependent proliferation and ERα transcriptional activity [2] [6]. |
| A1AR cDNA | Gene overexpression tool | Used in ERα-negative cell lines to demonstrate A1AR-induced proliferation [2]. |
This preliminary investigation consolidates evidence that the adenosine A1 receptor is a significant node in the signaling network of breast cancer. Its dual role as a target and regulator of ERα action, coupled with its ability to suppress p53-mediated apoptosis, positions it as a compelling therapeutic target. The consistent pro-tumorigenic effects of receptor agonists and the anti-cancer effects of antagonists like DPCPX across multiple studies provide a strong rationale for targeting A1AR. Emerging strategies, such as the dual inhibition of A1AR and ornithine decarboxylase (ODC) with molecules like ODC-MPI-2, demonstrate enhanced efficacy and represent a promising avenue for combination therapy [7]. Future research should focus on validating these findings in more complex in vivo models and exploring the translational potential of A1AR-targeted agents, particularly in hormone-dependent breast cancers where the crosstalk with ERα signaling can be exploited for therapeutic benefit.
The A1R within the Immunosuppressive Tumor Microenvironment of Breast Cancer
The adenosine A1 receptor (A1R) is a high-affinity G protein-coupled receptor (GPCR) for adenosine, traditionally studied for its roles in the central nervous system and cardiac function [10] [11]. Within the context of the immunosuppressive tumor microenvironment (TME) of breast cancer, the role of A1R is less characterized compared to other adenosine receptors like A2A and A2B. However, emerging evidence suggests it contributes significantly to the intricate network of immune evasion mechanisms that hinder effective anti-tumor responses [12] [13]. This whitepaper synthesizes current knowledge on A1R expression, signaling, and function within the breast cancer TME, framing it as a critical component of the hypoxia-adenosine immunosuppressive axis and a potential target for therapeutic intervention. A preliminary investigation into A1R reveals a complex role that warrants deeper exploration to fully exploit the adenosine pathway for breast cancer immunotherapy.
Adenosine Metabolism and Signaling in the TME
The immunosuppressive adenosine gradient in the TME is generated through a tightly regulated process. Under conditions of cellular stress, such as hypoxia within the tumor, intracellular ATP is released into the extracellular space [14]. Pannexin 1 (PANX1) channels facilitate this release of ATP [14]. The ecto-enzymes CD39 (ENTPD1) and CD73 (NT5E) then sequentially hydrolyze exATP to AMP and finally to adenosine [12] [14]. In triple-negative breast cancer (TNBC), tumor-associated neutrophils (TANs) have been identified as a source of these adenosine-generating enzymes [14].
Once produced, extracellular adenosine exerts its effects by binding to one of four adenosine receptors (A1, A2A, A2B, A3) on target cells. These receptors are GPCRs with differing affinities for adenosine; A1R and A2AR are high-affinity receptors, while A2BR and A3R have lower affinity [15] [11]. The A1R signals primarily through Gi/o proteins, leading to the inhibition of adenylyl cyclase and a decrease in intracellular cAMP levels [15] [11].
Table 1: Adenosine Receptors in the Tumor Microenvironment
| Receptor | Gene | Adenosine Affinity | Primary Signaling | General Role in Cancer Immunity |
|---|---|---|---|---|
| A1 | ADORA1 | High (nM range) | Gi/o → ↓ cAMP [15] [11] | Less defined; potential role in immune cell regulation [13] |
| A2A | ADORA2A | High (nM range) | Gs → ↑ cAMP [15] | Potent immunosuppression; inhibits T-cell and NK cell function [12] |
| A2B | ADORA2B | Low (µM range) | Gs / Gq → ↑ cAMP / ↑ IP3, DAG [15] | Involved in angiogenesis, inflammation; contributes to immunosuppression [12] |
| A3 | ADORA3 | Low (µM range) | Gi/o → ↓ cAMP [15] | Context-dependent roles in promoting apoptosis or survival [12] |
The following diagram illustrates the core pathway of adenosine generation and its immunosuppressive action via the A1 receptor on immune cells in the breast cancer TME.
A1R-Mediated Immunosuppressive Mechanisms in Breast Cancer
While the A2A receptor is often the primary focus in adenosine-mediated immunosuppression, A1R signaling contributes to shaping an inhibitory immune landscape in breast cancer through several mechanisms.
Direct Modulation of Immune Cell Function
A1R is expressed on various innate and adaptive immune cells, including monocytes, macrophages, neutrophils, dendritic cells, and microglia [13]. Activation of A1R on these cells can influence their function within the TME. In the context of breast cancer, high expression of PANX1, which drives adenosine production, is associated with increased recruitment of tumor-associated neutrophils (TANs) [14]. These TANs, in turn, express high levels of CD39 and CD73, creating a feed-forward loop for adenosine production and further amplifying local immunosuppression [14]. A1R signaling in other immune cells can inhibit pro-inflammatory responses and potentially promote phenotypes that support tumor growth [13].
Neuronal-like Inhibition in the TME
In the central nervous system, A1R activation provides potent neuronal inhibition through two principal mechanisms: membrane hyperpolarization via activation of potassium (K+) channels, and suppression of synaptic transmission via inhibition of voltage-gated calcium channels (VGCCs) and synaptic vesicle release [11]. An analogous process may occur within the TME, where A1R signaling on immune cells like T cells could lead to functional hyperpolarization and inhibition of cytotoxic effector functions, such as the release of perforin and granzymes, thereby silencing the anti-tumor immune response.
Investigating A1R in Breast Cancer: Research Landscape
Expression and Prognostic Value
The expression and clinical significance of immune checkpoint genes in breast cancer subtypes are highly variable. A systematic investigation of 50 immune checkpoint genes found that the expression pattern of ADORA2A (A2AR) was not significantly correlated with overall survival in breast cancer patients [16]. However, comprehensive data specifically on A1R (ADORA1) mRNA or protein expression levels across breast cancer molecular subtypes (Luminal A, Luminal B, HER2+, TNBC) and its direct prognostic value remains an area requiring further elucidation. This gap highlights the preliminary nature of A1R investigation in breast cancer.
Research Reagent and Experimental Toolkit
Studying A1R's role requires a suite of specific pharmacological and molecular tools. The table below details key reagents for in vitro and in vivo A1R research.
Table 2: Key Research Reagents for A1R Investigation
| Reagent / Tool | Category | Function / Specificity | Example Compounds / Identifiers |
|---|---|---|---|
| Selective Antagonists | Small Molecule | Competitively blocks A1R to investigate its function | DPCPX, CPT (8-cyclopentyltheophylline) [12] [15] |
| Selective Agonists | Small Molecule | Activates A1R to probe downstream signaling and effects | CCPA, Tecadenoson, Selodenoson [15] |
| Allosteric Modulators | Small Molecule | Binds a separate site to fine-tune orthosteric ligand activity (PAM/NAM) [10] | T-62 (PAM, reached clinical trial for pain) [10] |
| Non-Selective Antagonists | Small Molecule | Blocks multiple adenosine receptors (A1, A2A) | Caffeine, Theophylline [15] |
| shRNA/siRNA | Molecular Tool | Knocks down A1R gene expression for loss-of-function studies | PANX1 shRNA (used in TNBC models) [14] |
Proposed Experimental Workflow for A1R Investigation
A comprehensive research plan to delineate the role of A1R in breast cancer is essential. The following diagram outlines a multi-faceted experimental workflow, from initial assessment to functional validation.
Detailed Methodologies for Key Experiments
A1R Expression Profiling via Immunohistochemistry (IHC)
- Protocol: Formalin-fixed paraffin-embedded (FFPE) breast cancer tissue sections are deparaffinized and rehydrated. Antigen retrieval is performed by heating slides in citrate buffer (pH 6.0) for 20 minutes. Endogenous peroxidase is blocked with 3% H₂O₂. Sections are incubated overnight at 4°C with a validated primary anti-A1R antibody. After washing, a species-matched secondary antibody conjugated to HRP is applied for 30-60 minutes at room temperature. Staining is visualized using 3,3'-Diaminobenzidine (DAB) chromogen, and slides are counterstained with hematoxylin [14].
- Analysis: Staining is evaluated using a composite score based on intensity (0: negative, 1: weak, 2: positive, 3: strong) and frequency (1: 0-10%, 2: 11-30%, 3: 31-50%, 4: 51-75%, 5: 76-100%). The final score is the product of intensity and frequency. Samples can be dichotomized into A1R-high and A1R-low groups based on the median score [14].
Extracellular Adenosine (exADO) Quantification Assay
- Protocol: Breast cancer cell lines (e.g., MDA-MB-231, HCC-1937 for TNBC; MCF-7 for Luminal A) or single-cell suspensions from digested tumor specimens are plated in 24-well plates (2 x 10⁴ cells/well). After adherence, the culture medium is replaced with a fresh, serum-free medium. Cells are incubated for a predetermined time (e.g., 2-4 hours) under normoxic or hypoxic (1% O₂) conditions to mimic the TME. The cell culture supernatant is then collected and centrifuged at 4°C to remove any cellular debris [14].
- Analysis: The cleared supernatant is analyzed for adenosine concentration using a commercial competitive ELISA kit or liquid chromatography-mass spectrometry (LC-MS), following the manufacturer's protocol. Values are normalized to total cellular protein content.
T-cell Co-culture and Functional Assay
- Protocol: Peripheral blood mononuclear cells (PBMCs) are isolated from healthy donors. CD8+ T cells are purified using magnetic-activated cell sorting (MACS). Target breast cancer cells are co-cultured with activated CD8+ T cells at a predetermined ratio (e.g., 1:5) in the presence of an A1R-selective agonist (e.g., CCPA, 100 nM), an antagonist (e.g., DPCPX, 1 µM), or a vehicle control.
- Readouts:
- Cytokine Production: After 24-48 hours, supernatant is collected, and IFN-γ secretion is quantified by ELISA [17].
- T-cell Proliferation: CFSE-labeled T cells are used. After 3-5 days of co-culture, proliferation is assessed by flow cytometry based on CFSE dilution.
- Cytotoxicity: Real-time cytotoxicity is measured using platforms like Incucyte with caspase-based apoptosis assays.
Therapeutic Targeting of A1R and Future Directions
Targeting the adenosine pathway has emerged as a promising strategy in cancer immunotherapy. While most efforts have focused on A2AR antagonism or inhibition of CD39/CD73, A1R presents a complementary target [12].
Pharmacological Antagonism: Small-molecule A1R antagonists like DPCPX are valuable research tools. However, their therapeutic application may be limited by on-target side effects, particularly in the central nervous system and cardiovascular system, given A1R's physiological roles in reducing heart rate and promoting sedation [12] [10].
Allosteric Modulation: A promising approach to overcome the limitations of orthosteric drugs is the development of allosteric modulators. Positive allosteric modulators (PAMs) can enhance the effect of endogenous adenosine in a context-dependent manner, while negative allosteric modulators (NAMs) can inhibit it. Allosteric ligands can offer superior receptor subtype selectivity and a "ceiling effect" that may spare fundamental physiological signaling, potentially resulting in a safer therapeutic window [10]. The A1R PAM T-62 has previously reached clinical trials for neuropathic pain, demonstrating the clinical translatability of this mechanism [10].
Combination Therapies: Given the redundancy in immunosuppressive pathways, targeting A1R will likely be most effective in combination with other agents. Logical combinations include:
- Immune Checkpoint Inhibitors: Anti-PD-1/PD-L1 antibodies [18].
- A2AR Antagonists: To achieve broad-spectrum adenosine receptor blockade.
- CD39/CD73 Inhibitors: To reduce the production of the ligand (adenosine) itself [12].
In conclusion, the A1R represents a nuanced and underexplored component of the immunosuppressive network in breast cancer. A preliminary investigation confirms its presence and suggests mechanisms by which it contributes to immune evasion. Future research must systematically define its expression, function, and therapeutic potential using the detailed experimental frameworks and tools outlined herein.
Expression Patterns and Distribution of A1R in Breast Cancer Tissues
The adenosine A1 receptor (A1R, also designated as Adora1) has emerged as a significant molecular player in breast cancer pathogenesis, representing a promising target for therapeutic intervention. Within the context of breast cancer biology, A1R demonstrates a dual regulatory role, functioning both as a target and regulator of estrogen receptor-alpha (ERα) action [6]. This receptor mediates the proliferative effects of estradiol (E2), establishing a critical link between hormonal signaling and purinergic receptor pathways in cancer progression. The therapeutic potential of A1R inhibition is supported by experimental evidence demonstrating that selective A1R antagonists can significantly reduce breast cancer cell proliferation [6]. Recent advances in bioinformatics and computational chemistry have further validated A1R as a key candidate for targeted breast cancer treatment, with studies employing molecular docking and dynamics simulations to design compounds with high binding affinity for this receptor [9] [19]. This whitepaper comprehensively examines the expression patterns, distribution, and functional significance of A1R in breast cancer tissues, providing researchers with essential methodological frameworks for its investigation.
Biological Significance of A1R in Breast Cancer Progression
A1R participates in a feed-forward loop with estradiol and ERα that significantly favors breast cancer growth [6]. In ERα-positive breast cancer cells, estradiol upregulates A1R messenger RNA (mRNA) and protein levels, an effect that can be reversed by the E2 antagonist ICI 182,780 [6]. This regulatory relationship positions A1R as a crucial intermediary in hormonal signaling pathways that drive tumor proliferation.
The functional importance of A1R in breast cancer cell growth has been validated through multiple experimental approaches. Genetic ablation of A1R in ERα-positive cells significantly reduces both basal and E2-dependent proliferation, while A1R overexpression in ERα-negative cell lines induces proliferative activity [6]. Similarly, pharmacological inhibition using the selective A1R antagonist DPCPX effectively reduces proliferation, confirming A1R's role as a mediator of E2/ERα-dependent breast cancer growth [6].
Intriguingly, A1R ablation decreases both mRNA and protein levels of ERα and consequently diminishes estrogen-responsive element-dependent ERα transcriptional activity [6]. This regulatory effect extends to the binding activity of ERα to promoter regions of target genes such as TFF1, resulting in reduced TFF1 promoter activity and mRNA levels [6]. These findings collectively establish that A1R is required for the full transcriptional activity of ERα upon E2 stimulation, highlighting its fundamental role in hormone-responsive breast cancer signaling networks.
Quantitative Expression Profiles and Distribution Patterns
Expression Levels in Breast Cancer Models
Table 1: A1R Expression and Functional Assays in Breast Cancer Models
| Experimental Model | Expression/Effect | Measurement Method | Functional Outcome |
|---|---|---|---|
| ERα+ breast cancer cells | Upregulated by E2 | mRNA and protein analysis | Enhanced proliferation |
| A1R ablation models | Reduced ERα expression | mRNA/protein measurement | Decreased E2-dependent proliferation |
| A1R antagonist treatment | Inhibited proliferation | Cell viability assays | Reduced cancer growth |
| Molecular docking | Stable binding confirmed | LibDock Score: 148.673 [19] | Rational drug design |
The expression of A1R demonstrates cell-type specificity within breast cancer models. In ERα-positive cells, A1R expression is significantly enhanced by estradiol stimulation, creating a positive feedback loop that amplifies proliferative signaling [6]. This regulatory relationship is particularly relevant in luminal breast cancer subtypes, where hormonal signaling drives tumor progression.
Experimental evidence indicates that A1R targeting generates significant anti-proliferative effects across multiple breast cancer models. Small interference RNA-mediated ablation of A1R consistently reduces proliferative rates, while selective A1R antagonists demonstrate dose-dependent inhibition of cancer growth [6]. The stability of A1R-compound interactions has been confirmed through molecular dynamics simulations, with Compound 5 exhibiting particularly stable binding to the human adenosine A1 receptor-Gi2 protein complex (PDB ID: 7LD3) [9] [19].
Tissue Distribution Patterns
Table 2: A1R Distribution in Breast Tissue Compartments
| Tissue Compartment | A1R Expression Level | Association with Tumor Features | Methodological Approach |
|---|---|---|---|
| Tumor epithelium | Variable | Correlates with proliferation | IHC, mRNA analysis |
| Stromal compartment | Present | Microenvironment modulation | Immunohistochemistry |
| ERα+ tumors | Elevated | Hormone response | Comparative expression analysis |
| Invasive regions | Context-dependent | Potential invasion role | Spatial transcriptomics |
The distribution of A1R across breast tissue compartments reveals complex patterning that reflects its multifaceted role in cancer progression. While comprehensive immunohistochemical studies specifically detailing A1R distribution patterns in human breast cancer tissues are limited in the available literature, its expression is known to correlate with functional hormone responsiveness [6].
The spatial organization of A1R expression likely influences therapeutic accessibility and drug targeting efficacy. Recent methodological advances in spatial transcriptomics could provide more detailed mapping of A1R distribution patterns across different breast cancer subtypes and tissue regions [20]. Such approaches would help elucidate whether A1R demonstrates preferential expression in specific topological niches within the tumor microenvironment.
Experimental Protocols for A1R Investigation
Molecular Docking and Dynamics Simulations
Protocol Objective: To evaluate binding stability between candidate compounds and the human adenosine A1 receptor-Gi2 protein complex (PDB ID: 7LD3) [9] [19].
Step 1: System Preparation
- Obtain the crystal structure of the human adenosine A1 receptor-Gi2 protein complex (PDB ID: 7LD3)
- Prepare the ligand library using Discovery Studio 2019 Client
- Perform docking simulations with CHARMM to refine ligand shapes and charge distribution
Step 2: Molecular Docking
- Analyze binding interactions between compounds and A1R
- Select best poses based on LibDock scores
- Filter targets with scores exceeding 130 for further analysis [19]
- Compound 5 demonstrated a high LibDock score of 148.673 in studies [19]
Step 3: Molecular Dynamics (MD) Simulations
- Conduct MD simulations using GROMACS 2020.3
- Optimize protein structures with AMBER99SB-ILDN force field
- Model water molecules with TIP3P model
- Calculate ligand charges and generate GAFF force field-compatible files using ACPYPE
- Employ cubic boxes with minimum atom-box boundary distance of 0.8 nm
- Hydrate system with SOL water at 1000 g/L density
- Achieve electrical neutrality by replacing solvent water with chloride ions
- Perform initial energy minimization to relax the system
- Conduct 150 ps restrained MD simulation at 298.15 K
- Run unrestricted MD simulations with time step of 0.002 ps for 15 ns, maintaining isothermal-isobaric conditions at 298.15 K and 1 bar pressure [9]
Step 4: Trajectory Analysis
- Analyze motion trajectory of molecule interacting with target using VMD 1.9.3 software
- Capture data every 200 frames from initial to 8220th frame
- Document molecular dynamics throughout timeframe to understand binding process [9]
Functional Validation in Cellular Models
Protocol Objective: To evaluate the functional consequences of A1R modulation in breast cancer cells.
Step 1: Genetic Manipulation
- For A1R ablation: Utilize small interference RNA (siRNA) targeting A1R mRNA in ERα-positive cells
- For overexpression: Introduce A1R expression vector into ERα-negative cell lines
- Validate manipulation efficiency through quantitative RT-PCR and Western blotting
Step 2: Proliferation Assays
- Assess basal and E2-dependent proliferation rates
- Employ cell counting assays or metabolic activity measures (e.g., MTT, WST-1)
- Include selective A1R antagonist (DPCPX) treatment conditions
- Determine IC50 values for inhibitory compounds (e.g., Molecule 10 demonstrated IC50 = 0.032 µM against MCF-7 cells) [9] [19]
Step 3: Transcriptional Activity Assessment
- Measure ERα mRNA and protein levels following A1R manipulation
- Evaluate estrogen-responsive element-dependent ERα transcriptional activity using reporter assays
- Assess binding activity of ERα to target gene promoters (e.g., TFF1)
- Quantify downstream target gene expression (e.g., TFF1 mRNA levels) [6]
A1R Signaling Pathways in Breast Cancer
The adenosine A1 receptor participates in a complex signaling network that intersects with established oncogenic pathways in breast cancer. A1R activation triggers intracellular responses primarily through G-protein coupled receptor mechanisms, influencing key cellular processes including proliferation, survival, and gene expression.
Figure 1: A1R-ERα Feed-Forward Signaling Loop. This diagram illustrates the reciprocal regulatory relationship between A1R and ERα that promotes breast cancer growth. Estradiol binding to ERα promotes A1R expression, while A1R signaling stabilizes ERα and enhances its transcriptional activity, creating an amplification loop.
The cross-talk mechanism between A1R and ERα represents a clinically significant pathway in hormone-responsive breast cancers. This interaction creates a therapeutic opportunity for dual targeting approaches that simultaneously disrupt hormonal and purinergic signaling axes.
Research Reagent Solutions for A1R Investigation
Table 3: Essential Research Reagents for A1R Studies
| Reagent/Category | Specific Examples | Research Application | Experimental Function |
|---|---|---|---|
| Cell Lines | MCF-7 (ER+), MDA-MB-231 (ER-) | In vitro models | Cellular proliferation, gene expression studies |
| A1R Agonists | N/A | Pathway activation | Study A1R-mediated effects |
| A1R Antagonists | DPCPX | Pharmacological inhibition | Validate A1R-dependent phenotypes |
| siRNA/shRNA | A1R-targeting sequences | Genetic ablation | Determine A1R necessity in processes |
| Antibodies | Anti-A1R, Anti-ERα | Protein detection | IHC, Western blot, immunofluorescence |
| Molecular Biology | qPCR primers, reporter constructs | Expression and promoter studies | Quantify mRNA, measure activity |
| Computational Tools | Discovery Studio, GROMACS, VMD | Molecular modeling | Docking, dynamics simulations, visualization |
The selection of appropriate research tools is critical for rigorous investigation of A1R in breast cancer models. The MCF-7 cell line (ER+) is particularly relevant for studying A1R-ERα interactions, while MDA-MB-231 (ER-) cells serve as useful comparators [9]. The selective A1R antagonist DPCPX has demonstrated efficacy in functional studies, providing a pharmacological tool for target validation [6].
For computational studies, the human adenosine A1 receptor-Gi2 protein complex (PDB ID: 7LD3) serves as a key structural template for molecular docking and dynamics simulations [9] [19]. The integration of these computational approaches with experimental validation creates a powerful framework for advancing A1R-targeted therapeutic development.
The expression patterns and distribution of A1R in breast cancer tissues reveal a receptor with significant pathophysiological importance and therapeutic potential. As both a regulator and target of ERα action, A1R occupies a strategic position in hormone-responsive breast cancer signaling networks. The development of selective A1R antagonists represents a promising avenue for therapeutic intervention, particularly in combination with established endocrine therapies.
Future research directions should include comprehensive tissue mapping of A1R expression across breast cancer subtypes using advanced spatial transcriptomics and proteomics approaches. Additionally, the exploration of A1R's role in the tumor microenvironment may reveal novel mechanisms of cancer-stroma communication that influence disease progression and treatment response. The continued investigation of A1R in breast cancer will likely yield important insights into cancer biology and contribute to the development of more effective, targeted therapeutic strategies.
The Dual Role of Adenosine Receptors in Tumor Promotion and Suppression
Adenosine receptors (ARs), comprising four subtypes—A1, A2A, A2B, and A3—are G protein-coupled receptors (GPCRs) that serve as the endogenous receptors for adenosine [21]. These receptors play pivotal roles in numerous physiological and pathological processes, with their significance in cancer biology increasingly recognized. The tumor microenvironment (TME) is characterized by elevated adenosine concentrations, often reaching micromolar levels due to hypoxia and extensive ATP breakdown from necrotic cells [22] [21]. This adenosine-rich milieu engages adenosine receptors in complex signaling networks that exert both tumor-promoting and tumor-suppressive effects, creating a dynamic regulatory system that influences cancer progression, immune evasion, and therapeutic responses.
The dual nature of adenosine receptor signaling in cancer presents a fascinating paradox in oncology research. While certain receptor subtypes and their signaling pathways appear to drive tumor growth and immunosuppression, others can trigger cytostatic and apoptotic pathways in malignant cells [22]. This review explores the complex interplay of adenosine receptor subtypes in cancer, with particular emphasis on the adenosine A1 receptor (A1AR) in breast cancer, integrating current understanding of their mechanisms, experimental approaches for their investigation, and potential therapeutic implications.
Adenosine Receptor Subtypes: Structure, Signaling, and Expression
Molecular Characteristics and Signaling Pathways
Adenosine receptors belong to the class A family of rhodopsin-like GPCRs, each possessing seven transmembrane domains with an extracellular N-terminus and intracellular C-terminus [22]. Despite their structural similarities, these receptors couple to different G proteins and activate distinct intracellular signaling cascades:
A1 Adenosine Receptor (A1AR): The A1 receptor primarily couples to pertussis toxin-sensitive Gαi and Gαo proteins [23]. Upon activation, it inhibits adenylate cyclase, reducing intracellular cAMP levels [24] [25] [23]. Additionally, A1AR modulates various ion channels, inhibiting N-, P-, and Q-type calcium channels while activating several potassium channels [25] [23]. The receptor also activates phospholipase C (PLC) via Gβγ subunits, leading to inositol trisphosphate (IP3) production and intracellular calcium mobilization [23].
A2A Adenosine Receptor (A2AAR): In contrast to A1AR, A2AAR couples to Gαs proteins, stimulating adenylate cyclase and increasing cAMP formation [24] [22]. This receptor is highly expressed in immune cells, particularly neutrophils and regulatory T cells, where it contributes to immunosuppressive pathways [21].
A2B Adenosine Receptor (A2BAR): Like A2AAR, A2BAR stimulates adenylate cyclase through Gαs coupling but has lower affinity for adenosine [24]. It is expressed in various cell types, including fibroblasts, endothelial cells, and immune cells such as bone marrow cells and basophils [21].
A3 Adenosine Receptor (A3AR): A3AR primarily couples to Gαi proteins, inhibiting adenylate cyclase and reducing cAMP levels [22]. It can also interact with Gq and Go proteins, activating phospholipase C and modulating calcium mobilization [22]. A3AR is overexpressed in numerous cancer types and demonstrates a unique dual role in regulating cell proliferation and death [22].
Table 1: Adenosine Receptor Subtypes and Their Key Signaling Pathways
| Receptor Subtype | G Protein Coupling | Primary Signaling Effects | Adenosine Affinity |
|---|---|---|---|
| A1AR | Gi/o | Inhibits adenylate cyclase ↓ cAMP, activates PLC, modulates K+ and Ca2+ channels | High (nanomolar) |
| A2AAR | Gs | Stimulates adenylate cyclase ↑ cAMP | High (nanomolar) |
| A2BAR | Gs | Stimulates adenylate cyclase ↑ cAMP | Low (micromolar) |
| A3AR | Gi/o | Inhibits adenylate cyclase ↓ cAMP, activates PLC, modulates MAPK/PI3K | Intermediate |
Expression Patterns in Normal and Neoplastic Tissues
Adenosine receptor expression varies significantly across tissues and cell types, influencing their specific roles in physiological and pathological conditions. The A2AAR protein is expressed predominantly in the colon, caudate nucleus, appendix, cerebellum, kidney, and bone marrow [21]. In immune cells, A2AAR levels are higher in neutrophils and regulatory T cells (Treg cells) [21]. A1AR demonstrates widespread distribution throughout the body, with particularly high expression in the cerebral cortex, hippocampus, cerebellum, thalamus, basal ganglia, brainstem, and spinal cord [23]. A1AR protein shows tissue-specific localization in Müller glial cells, oligodendrocyte precursor cells, oligodendrocytes, and excitatory neurons [21].
In cancer tissues, adenosine receptor expression often undergoes significant alterations. A3AR is overexpressed in various malignancies, including melanoma, breast, prostate, liver, pancreatic, and lung cancers, as well as lymphoma, glioblastoma, and malignant pleural mesothelioma [22]. In breast cancer specifically, A1AR has been identified as both a target and regulator of estrogen receptor α (ERα) action, mediating the proliferative effects of estradiol [26]. The A1 receptor is upregulated by estradiol in ERα-positive breast cancer cells, establishing a feed-forward loop that promotes cancer growth [26].
The Dual Nature of Adenosine Receptors in Tumor Biology
Tumor-Promoting Functions
Adenosine receptors contribute to tumor progression through multiple mechanisms that enhance cancer cell survival, proliferation, and immune evasion:
Immunosuppression: The adenosine-rich TME engages A2AAR and A2BAR on immune cells, creating potent immunosuppressive signals [21]. A2AAR activation inhibits the activity of various immune cells, including T cells and natural killer (NK) cells, thereby dampening antitumor immunity [21]. Dual A2A/A2B antagonists like M1069 have demonstrated superior suppression of protumorigenic cytokines and enhanced T-cell stimulatory activity compared to selective A2A antagonists [27].
Angiogenesis Promotion: Adenosine signaling stimulates new blood vessel formation to support tumor growth. A2BAR activation promotes the secretion of pro-angiogenic factors such as vascular endothelial growth factor (VEGF), CXCL12, and FGF2 [21]. In melanoma models, A2BAR inhibition reduced the number of cancer-associated fibroblasts (CAFs) expressing fibroblast activation protein (FAP) and FGF-2, consequently impairing tumor vascularization [21].
Proliferation and Survival Signaling: In breast cancer, A1AR mediates the proliferative effects of estradiol and is required for full transcriptional activity of ERα [26]. Ablation of A1AR in ERα-positive cells reduces basal and estradiol-dependent proliferation, while A1AR overexpression in ERα-negative cell lines induces proliferation [26]. Similarly, A3AR activation in certain cancer contexts leads to decreased cAMP levels, resulting in reduced phosphorylated PKB/Akt and PKA activity, thereby dysregulating Wnt signaling and promoting tumorigenesis [22].
Tumor-Suppressive Functions
Paradoxically, adenosine receptors can also trigger pathways that inhibit tumor growth and survival:
Direct Antiproliferative and Pro-apoptotic Effects: A3AR demonstrates a concentration-dependent dual functionality. At high (micromolar) concentrations, selective synthetic A3AR agonists exhibit pro-apoptotic effects in both normal and tumor cells [22]. In leukemia and melanoma models, high adenosine levels have demonstrated pro-apoptotic effects [22]. Low adenosine concentrations (<25 nM) have also been reported to inhibit tumor growth in certain contexts [22].
Immunomodulation: While generally immunosuppressive, adenosine receptors can under specific conditions modulate immune responses in ways that may limit tumor progression. A3AR activation inhibits chemotaxis, degranulation, and superoxide anion generation in eosinophils [22]. In monocytes and macrophages, A3AR inhibits TNF-α release through the NF-κB signal transduction pathway [22].
Cell Cycle Regulation: A3AR activation can inhibit tumor growth by regulating the Wnt pathway [22]. Through modulation of glycogen synthase kinase-3β (GSK-3β), A3AR signaling can lead to down-regulation of β-catenin and cyclin D1, key regulators of cell cycle progression [22].
Table 2: Dual Roles of Adenosine Receptors in Cancer Processes
| Cancer Process | Tumor-Promoting Effects | Tumor-Suppressive Effects |
|---|---|---|
| Immune Regulation | A2A/A2B: Suppress T-cell and NK cell activity [27] [21] | A3: Inhibits TNF-α release in macrophages [22] |
| Cell Proliferation | A1: Mediates E2/ERα-dependent proliferation in breast cancer [26] | A3: High agonist concentrations induce apoptosis [22] |
| Angiogenesis | A2B: Promotes VEGF, CXCL12, FGF2 secretion [21] | Low adenosine (<25 nM) inhibits tumor growth [22] |
| Cell Signaling | A3: Dysregulates Wnt signaling via GSK-3β [22] | A3: Regulates Wnt pathway to inhibit growth [22] |
Preliminary Investigation of Adenosine A1 Receptor in Breast Cancer Research
A1AR as a Regulator of Estrogen Receptor α Signaling
The adenosine A1 receptor plays a particularly significant role in hormone-dependent breast cancer, where it functions as both a target and regulator of estrogen receptor α (ERα) action [26]. In ERα-positive breast cancer cells, estradiol (E2) upregulates A1AR mRNA and protein levels, an effect that is reversed by the estrogen antagonist ICI 182,780 [26]. This establishes A1AR as a direct target of ERα signaling. Intriguingly, A1AR also regulates ERα expression and activity, creating a short feed-forward loop that favors breast cancer growth [26].
RNA interference-mediated ablation of A1AR in ERα-positive cells reduces both basal and E2-dependent proliferation, while A1AR overexpression in ERα-negative cell lines induces proliferation [26]. The selective A1AR antagonist, DPCPX, similarly reduces proliferation, confirming A1AR as a mediator of E2/ERα-dependent breast cancer growth [26]. Mechanistically, A1AR ablation decreases both mRNA and protein levels of ERα and consequently reduces estrogen-responsive element-dependent ERα transcriptional activity [26]. Furthermore, A1AR ablation decreases binding of ERα to the promoter of its target gene TFF1, leading to reduced TFF1 promoter activity and mRNA levels [26].
Experimental Approaches for A1AR Investigation in Breast Cancer
Computational and Bioinformatics Methods
Recent studies have employed integrated bioinformatics and computational chemistry approaches to identify critical therapeutic targets and design potent antitumor compounds targeting adenosine receptors in breast cancer [9]. These methodologies include:
Target Screening and Intersection Analysis: Initial screening and target intersection analysis identified the adenosine A1 receptor as a key candidate for breast cancer treatment [9]. Shared targets across multiple anticancer compounds were identified using online tools such as Venny, revealing common pathways in MDA-MB and MCF-7 breast cancer cell lines [9].
Molecular Docking Simulations: Molecular docking with CHARMM force fields has been used to refine ligand shapes and charge distribution while analyzing binding interactions between compounds and the human adenosine A1 receptor-Gi2 protein complex (PDB ID: 7LD3) [9]. Targets with LibDock scores over 130 were selected for further investigation, providing insights into binding mechanisms [9].
Molecular Dynamics (MD) Simulations: MD simulations using GROMACS analyzed protein-ligand binding dynamics with optimized protein structures using the AMBER99SB-ILDN force field [9]. Water molecules were modeled with the TIP3P model, and ligand charges were calculated using ACPYPE to generate GAFF force field-compatible files [9]. Simulations typically employed cubic boxes with minimum atom-box boundary distances of 0.8 nm, hydrated with SOL water at 1000 g/L density, with chloride ions replacing solvent water for electrical neutrality [9].
Pharmacophore Modeling: Construction of pharmacophore models based on binding information guides virtual screening of additional compounds with activity against A1AR [9]. These models define key structural features influencing biological activity and facilitate rational drug design.
A1AR Signaling in Breast Cancer
In Vitro Validation Methods
Experimental validation of computational findings involves comprehensive in vitro approaches:
Cell Culture Models: Estrogen receptor-positive (ER+) MCF-7 cell lines derived from human breast cancer tissue are widely used to investigate estrogen dependency and evaluate therapies targeting estrogen signaling pathways [9]. In contrast, MDA-MB cell lines, characterized by lack of estrogen receptor expression (ER-), are employed for studying aggressive breast cancer behaviors such as metastasis and drug resistance [9].
Biological Evaluation: Designed compounds are synthesized and evaluated for antitumor activity using MCF-7 breast cancer cells [9]. For instance, Molecule 10, rationally designed based on pharmacophore models, demonstrated potent antitumor activity with an IC50 value of 0.032 µM, significantly outperforming the positive control 5-FU (IC50 = 0.45 µM) [9].
Mechanistic Studies: The distribution of dynamic binding positions of compounds with targets is analyzed using software such as VMD 1.9.3 [9]. Comprehensive analysis of motion trajectories from molecular dynamics simulations, typically spanning thousands of frames with data recorded at regular intervals, facilitates understanding of molecular binding processes, dynamic behavior, and potential intermediate states [9].
Research Reagent Solutions for A1AR Investigation
Table 3: Essential Research Reagents for Adenosine A1 Receptor Studies
| Reagent/Category | Specific Examples | Function/Application |
|---|---|---|
| Cell Lines | MCF-7 (ER+), MDA-MB (ER-) | Model systems for studying A1AR in different breast cancer subtypes [9] |
| A1AR Agonists | CCPA, N6-Cyclopentyladenosine, Capadenoson (BAY68-4986) | Selective activation of A1AR signaling pathways [25] |
| A1AR Antagonists | DPCPX, Rolofylline, CVT-124 | Inhibit A1AR function; study loss-of-function effects [26] [28] |
| Computational Tools | GROMACS, VMD, Discovery Studio | Molecular docking, dynamics simulations, visualization [9] |
| Antibodies | A1AR-specific antibodies | Detect protein expression and localization via Western blot, IHC [26] |
| Gene Silencing Tools | siRNA against ADORA1 | Knockdown A1AR expression for functional studies [26] |
Therapeutic Implications and Future Directions
Targeting Adenosine Receptors in Cancer Therapy
The dual nature of adenosine receptors in tumor biology presents both challenges and opportunities for therapeutic intervention. Several strategies have emerged:
Receptor Antagonists: Selective A1AR antagonists such as DPCPX have demonstrated efficacy in reducing breast cancer proliferation [26]. Dual A2A/A2B antagonists like M1069 counteract immunosuppressive mechanisms of adenosine and reduce tumor growth in vivo, particularly in adenosine-high/CD73hi tumor models such as 4T1 breast tumors [27].
Receptor Agonists: Interestingly, A3AR agonists demonstrate concentration-dependent antitumor effects, with high (micromolar) concentrations triggering apoptotic pathways in cancer cells [22]. This suggests that context-specific agonist administration might harness the tumor-suppressive potential of adenosine receptors.
Combination Therapies: Adenosine receptor antagonists enhance the efficacy of existing treatments. M1069 improved the antitumor activity of bintrafusp alfa (BA) and cisplatin in syngeneic adenosinehi/CD73hi 4T1 breast tumor models [27]. Similarly, combining adenosine receptor inhibition with immunotherapy may overcome resistance mechanisms in the TME [21].
Experimental Workflow for A1AR-Targeted Drug Discovery
Drug Discovery Workflow
Challenges and Future Perspectives
Despite promising developments, several challenges remain in targeting adenosine receptors for cancer therapy. The dual nature of these receptors, particularly A3AR and A1AR, necessitates careful context-specific therapeutic approaches. Tissue-specific distribution and expression patterns of adenosine receptors complicate systemic administration of targeted therapies. Furthermore, the complex interplay between different adenosine receptor subtypes in the TME may lead to compensatory mechanisms and treatment resistance.
Future research should focus on developing more selective modulators of adenosine receptors, optimizing combination strategies with conventional therapies and immunotherapy, and identifying biomarkers that predict response to adenosine-targeted therapies. Additionally, advanced drug delivery systems that enable tissue-specific targeting may enhance therapeutic efficacy while minimizing off-target effects.
In conclusion, adenosine receptors, particularly the A1 receptor in breast cancer, represent promising therapeutic targets whose dual nature in tumor promotion and suppression reflects the complexity of cancer biology. Through integrated computational, experimental, and therapeutic approaches, targeting these receptors may yield innovative strategies for cancer treatment.
The adenosine A1 receptor (A1R), a class A G protein-coupled receptor (GPCR), has emerged as a significant regulator in the tumor microenvironment with particularly intriguing implications for breast cancer pathogenesis. Within the context of a broader thesis on the preliminary investigation of A1R in breast cancer research, this technical review synthesizes current understanding of how A1R signaling impacts three fundamental cancer hallmarks: cellular proliferation, apoptotic evasion, and angiogenesis. Adenosine, the endogenous orthosteric ligand for A1R, accumulates to high concentrations in hypoxic tumor microenvironments through the enzymatic conversion of extracellular ATP by CD39 and CD73 [10]. This review examines the mechanistic basis for A1R-mediated oncogenic signaling and its potential as a therapeutic target, providing detailed experimental frameworks for researchers investigating GPCR biology in cancer systems.
A1R Signaling in Breast Cancer Proliferation
Molecular Mechanisms Linking A1R to Proliferative Signaling
The adenosine A1 receptor exhibits a complex functional relationship with estrogen receptor alpha (ERα) signaling in breast cancer, forming a feed-forward loop that drives cellular proliferation. Research has demonstrated that A1R functions as both a target and regulator of ERα action [6]. In ERα-positive breast cancer cells, estradiol (E2) significantly upregulates A1R mRNA and protein expression, an effect that is reversible by the ER antagonist ICI 182,780 [6]. This establishes A1R as a direct transcriptional target of the ligand-activated ERα.
The proliferative influence of A1R signaling has been validated through multiple experimental approaches. Small interfering RNA (siRNA)-mediated ablation of Adora1 (the gene encoding A1R) in ERα-positive breast cancer cells substantially reduces both basal and E2-dependent proliferation [6]. Conversely, A1R overexpression in ERα-negative cell lines induces proliferative capacity, while treatment with the selective A1R antagonist DPCPX (8-cyclopentyl-1,3-dipropylxanthine) effectively suppresses proliferation in A1R-expressing systems [6]. Intriguingly, A1R ablation decreases both mRNA and protein levels of ERα itself, consequently diminishing estrogen-responsive element (ERE)-dependent transcriptional activity and expression of endogenous ERα target genes such as TFF1 [6].
Table 1: Experimental Evidence for A1R-Mediated Proliferation in Breast Cancer
| Experimental System | Intervention | Key Findings | Citation |
|---|---|---|---|
| MCF-7 ERα+ cells | siRNA Adora1 ablation | Reduced basal and E2-dependent proliferation | [6] |
| ERα- cell line | A1R overexpression | Induced proliferation | [6] |
| ERα+ breast cancer cells | DPCPX (A1R antagonist) | Decreased proliferation | [6] |
| MCF-7 cells | Compound 10 (novel A1R-targeting molecule) | IC50 = 0.032 µM (vs. 0.45 µM for 5-FU control) | [9] |
Experimental Protocols for Investigating A1R-Mediated Proliferation
Protocol 1: siRNA-Mediated A1R Ablation and Proliferation Assessment
- Cell Lines: MCF-7 (ERα-positive breast cancer cells)
- A1R Knockdown: Transfect with 50-100 nM validated Adora1-specific siRNA using appropriate transfection reagent
- Controls: Non-targeting siRNA and mock transfection controls
- Proliferation Assay: Assess cell viability at 24, 48, 72, and 96h post-transfection using MTT or CCK-8 assays
- Hormone Stimulation: Treat with 10 nM E2 (with/without 100 nM ICI 182,780) 24h post-transfection
- Validation: Confirm A1R knockdown via qPCR (mRNA) and Western blot (protein) [6]
Protocol 2: A1R Antagonism Studies
- Compound: DPCPX (selective A1R antagonist) at concentrations ranging from 10 nM to 10 µM
- Treatment Duration: 24-96 hours
- Endpoint Measurements: Cell counting, 3H-thymidine incorporation, or flow cytometric analysis of cell cycle distribution [6]
A1R Modulation of Apoptotic Pathways
A1R Signaling and Cell Death Regulation
The adenosine A1 receptor demonstrates context-dependent effects on apoptotic pathways in breast cancer systems. In MCF-7 breast cancer cells, treatment with an A1R agonist modulates the expression of key apoptosis regulators including p53 and multiple caspases [29]. Specifically, studies have documented that A1R agonist treatment influences the expression levels of caspase-3, -8, and -9, core components of the apoptotic machinery [29]. These findings position A1R as a potential regulator of programmed cell death decisions in breast cancer cells, though the precise mechanisms appear to vary based on cellular context and receptor expression levels.
The interplay between A1R and apoptotic regulation extends to other cancer types, suggesting potential conserved mechanisms. In oral squamous cell carcinoma, progression from normal mucosa to dysplasia and invasive carcinoma is accompanied by significant increases in apoptotic indices alongside parallel increases in proliferation and angiogenesis [30]. This coordinated regulation suggests that A1R signaling may function within broader networks that balance cell death and survival decisions in epithelial malignancies.
Table 2: A1R-Mediated Effects on Apoptosis Regulators
| Apoptosis Component | A1R-Mediated Effect | Experimental System | Functional Outcome |
|---|---|---|---|
| p53 expression | Modulated by A1R agonist | MCF-7 cells | Altered transcription of p53 target genes |
| Caspase-3, -8, -9 | Expression changes | MCF-7 cells | Modified execution of apoptotic programs |
| Apoptotic Index | Increases with disease progression | Oral tissue carcinogenesis | Balanced with proliferation increases |
Experimental Protocols for Apoptosis Analysis
Protocol 3: A1R Modulation and Apoptosis Assessment
- Cell Preparation: Seed MCF-7 cells in 6-well plates (2×10^5 cells/well)
- A1R Modulation: Treat with A1R agonist (e.g., CCPA; 100 nM) or antagonist (DPCPX; 1 µM) for 24-48h
- Apoptosis Detection:
- Annexin V/PI Staining: Use commercial apoptosis detection kit per manufacturer's protocol
- Caspase Activity: Measure using fluorometric caspase activity assays
- Western Blotting: Analyze cleaved PARP, caspases, and p53 expression
- Gene Expression: Quantify mRNA levels of p53 and caspase genes via RT-qPCR [29]
Protocol 4: In Situ Apoptosis Detection in Tissue Models
- Tissue Preparation: Formalin-fixed, paraffin-embedded sections (5 µm thickness)
- Apoptosis Labeling: Employ TUNEL assay (in-situ end-labeling of DNA fragments)
- Visualization: Enzyme-based colorimetric detection or fluorescence labeling
- Quantification: Count apoptotic cells in 10 random high-power fields (400× magnification) [30]
A1R Involvement in Tumor Angiogenesis
Angiogenic Signaling Mediated by A1R
While direct evidence specifically linking A1R to angiogenesis in breast cancer requires further investigation, the broader context of adenosine receptor signaling supports a likely role in promoting tumor vascularization. In multiple cancer models, adenosine signaling has been demonstrated to stimulate angiogenesis through various mechanisms, primarily attributed to the A2A and A2B receptor subtypes [29]. These pro-angiogenic effects include upregulation of vascular endothelial growth factor (VEGF) and other angiogenic factors.
The methodology for assessing angiogenesis in relation to adenosine signaling has been established in other tissue contexts. In oral carcinogenesis, angiogenesis measurement typically involves immunostaining for endothelial cell markers followed by microvessel density quantification [30]. This approach could be readily adapted to breast cancer models to specifically elucidate A1R contributions. The significant increase in vascularity observed during progression from normal oral mucosa through dysplasia to invasive carcinoma demonstrates the importance of angiogenesis in epithelial cancer development [30].
Experimental Protocols for Angiogenesis Assessment
Protocol 5: Microvessel Density Quantification
- Tissue Staining: Perform immunohistochemistry on formalin-fixed paraffin-embedded sections using von Willebrand factor (vWF) or CD31 antibodies to highlight endothelial cells
- Microvessel Counting: Identify vascular hotspots at 100× magnification, then count microvessels in three 200× fields
- Quantification Criteria: Any brown-staining endothelial cell or cell cluster clearly separate from adjacent microvessels counts as a single microvessel
- Analysis: Calculate mean microvessel density ± standard deviation across samples [30]
Protocol 6: In Vitro Angiogenesis Assays
- Endothelial Cell Culture: Human umbilical vein endothelial cells (HUVECs) or other relevant endothelial cell types
- Conditioned Media Collection: Culture breast cancer cells with A1R modulators, collect conditioned media after 48h
- Tube Formation Assay: Seed endothelial cells on Matrigel-coated plates, treat with conditioned media, quantify tube formation after 4-16h
- Analysis: Measure total tube length, number of branches, and enclosed areas using image analysis software
Structural Insights and Therapeutic Targeting of A1R
A1R Allosteric Modulators as Therapeutic Opportunities
Recent structural biology advances have revealed novel approaches for targeting A1R with improved selectivity. The crystal structure of A1R bound to adenosine and the positive allosteric modulator MIPS521 ([2-amino-4-(3,5-bis(trifluoromethyl)phenyl)thiophen-3-yl)(4-chlorophenyl)methanone]) in complex with a Gi2 heterotrimer has been resolved [31]. This structure reveals an extrahelical lipid–detergent-facing allosteric binding pocket involving transmembrane helixes 1, 6 and 7, distinct from the orthosteric adenosine-binding site [31]. Molecular dynamics simulations and kinetic binding experiments indicate that MIPS521 stabilizes the adenosine–receptor–G protein complex, providing a mechanism for its positive allosteric modulation [31].
This structural insight enables structure-based drug design of non-opioid analgesic agents that are specific to disease contexts [31], and similar approaches could be applied to breast cancer therapeutics. Positive allosteric modulators of A1R represent a promising strategy because they enhance the receptor's response to endogenous adenosine only in tissues and disease states where adenosine concentrations are elevated, potentially offering improved therapeutic windows compared to orthosteric agonists [10].
Computational Approaches for A1R-Targeted Drug Discovery
Protocol 7: Molecular Docking and Dynamics for A1R Ligand Discovery
- Protein Preparation: Retrieve A1R structure (PDB: 7LD3), remove crystallographic waters, add hydrogen atoms, assign charges
- Ligand Preparation: Draw candidate compounds, optimize geometry, generate 3D conformers
- Molecular Docking: Use AutoDock Vina or similar software with binding pocket defined by allosteric site coordinates
- Molecular Dynamics: Run simulations using GROMACS with AMBER99SB-ILDN force field for proteins and GAFF for ligands
- Analysis: Calculate binding free energies, monitor conformational changes, identify key interacting residues [9]
Protocol 8: Pharmacophore-Based Virtual Screening
- Pharmacophore Generation: Define spatial features based on known active compounds (hydrogen bond donors/acceptors, hydrophobic regions, aromatic rings)
- Database Screening: Filter compound libraries (ZINC, PubChem) against pharmacophore model
- Compound Selection: Prioritize compounds with high fit values and favorable drug-like properties
- Experimental Validation: Test selected compounds in binding and functional assays [9]
Research Reagent Solutions
Table 3: Essential Research Reagents for Investigating A1R in Breast Cancer
| Reagent/Category | Specific Examples | Function/Application | Experimental Notes |
|---|---|---|---|
| A1R Agonists | CCPA, CPA, ADAC | Activate A1R signaling; study downstream effects | Use concentration range 1 nM-1 µM; monitor receptor internalization |
| A1R Antagonists | DPCPX, CPT | Inhibit A1R signaling; validate receptor-specific effects | DPCPX selective at concentrations <100 nM |
| Allosteric Modulators | MIPS521, T-62, TRR469 | Enhance (PAM) or inhibit (NAM) endogenous adenosine effects | Display probe dependence; test with multiple orthosteric ligands |
| Antibodies | Anti-A1R (various hosts), anti-vWF, anti-Ki-67 | Detect protein expression, localization; stain blood vessels, proliferating cells | Validate specificity using knockout controls or competing peptides |
| Cell Lines | MCF-7 (ERα+), MDA-MB-231 (ERα-) | Model different breast cancer subtypes | Monitor receptor expression across passages |
| siRNA/shRNA | Adora1-targeting sequences | Knockdown A1R expression; study loss-of-function | Include multiple distinct sequences to control for off-target effects |
| Computational Tools | GROMACS, AutoDock Vina, SwissTargetPrediction | Molecular dynamics, docking, target prediction | Use updated force fields; validate predictions experimentally |
Visualizing A1R Signaling and Research Approaches
A1R Signaling Pathway in Breast Cancer
Experimental Workflow for A1R Research
The adenosine A1 receptor represents a multifaceted regulator in breast cancer biology, simultaneously influencing proliferative signaling through integration with ERα pathways, modulating apoptotic responses, and potentially contributing to angiogenic processes. The experimental frameworks and technical approaches detailed in this review provide researchers with comprehensive methodologies for investigating A1R function in breast cancer models. The emerging structural understanding of A1R and development of allosteric modulators offer promising avenues for therapeutic intervention with potentially improved selectivity. Future research directions should include elucidating A1R interactions with other adenosine receptor subtypes in the tumor microenvironment, exploring A1R roles in different breast cancer molecular subtypes, and validating A1R-targeting strategies in more complex preclinical models that recapitulate the hypoxic, adenosine-rich tumor microenvironment.
Advanced Methodologies for A1R Target Screening and Compound Design in Breast Cancer
Integrated Bioinformatics and Computational Chemistry for A1R Target Identification
The preliminary investigation of the adenosine A1 receptor (A1R) within breast cancer research represents a promising frontier for therapeutic intervention. This technical guide delineates an integrated framework employing bioinformatics and computational chemistry methodologies for the identification and validation of A1R as a molecular target. We provide detailed protocols for structural analysis, molecular docking, dynamics simulations, and data integration, supplemented by structured tables of quantitative data and visual workflows. This approach aims to establish a foundational pipeline for researchers targeting A1R in the context of breast cancer pathogenesis and treatment.
G-protein-coupled receptors (GPCRs) like the adenosine A1 receptor are critical regulators of cellular signaling and represent a major class of drug targets. The A1R preferentially couples to inhibitory Gi/o heterotrimeric G proteins and has been implicated in numerous disease pathways [32]. Within breast cancer, a complex and prevalent health concern affecting millions worldwide, understanding receptor-mediated signaling pathways offers significant potential for therapeutic advancement [33]. Breast cancer encompasses heterogeneous molecular subtypes with varying receptor expressions (estrogen, progesterone, and HER2 receptors), with triple-negative breast cancer (TNBC) representing an aggressive subtype with limited treatment options and poor prognosis [34]. The investigation of alternative molecular targets like A1R is thus clinically imperative.
The endogenous agonist for A1R is adenosine, which plays a role in modulating pain pathways and has been investigated as a non-opioid analgesic target [35]. The structure of the adenosine-bound human A1R-Gi complex has been resolved, revealing critical molecular interactions at the orthosteric binding site mediated via transmembrane domains 1 and 2, and intracellular engagement with G proteins [32] [35]. This structural knowledge enables targeted investigation of A1R's role in breast cancer through computational means, integrating bioinformatic analyses of expression data with computational chemistry approaches for ligand discovery and optimization.
Bioinformatics Approaches for A1R Target Analysis
Transcriptomic Data Integration and Analysis
The analysis of A1R expression across breast cancer subtypes requires robust integration of transcriptomic data. RNA-sequencing (RNA-seq) data from public repositories like the NCBI Gene Expression Omnibus (GEO) often originate from different platforms (e.g., polyA-selected or rRNA-depleted libraries), creating challenges for comparative analysis.
Experimental Protocol: Transcriptomic Data Integration using GEDI R Package
- Data Import: Use the
ReadGEfunction from the GEDI package to import gene expression datasets from multiple sources (e.g., microarray and RNA-seq) [36]. - Reannotation and Merging: Execute the
GEDIfunction to automatically reannotate all gene identifiers to a common standard (e.g., official gene symbols) and merge the datasets into a single expression matrix [36]. - Batch Effect Correction: Apply the
BatchCorrectionfunction to remove non-biological technical variation between datasets using established algorithms like ComBat [36]. - Integration Verification: Utilize
VerifyGEDIto confirm successful data integration. This function employs principal component analysis (PCA) to visualize batch effect removal and uses a logistic regression model with forward stepwise feature selection to validate the integration [36].
For RNA-seq data specifically, gene-level expression estimates can be obtained using high-speed transcript quantification tools like Kallisto. A study on ovine macrophages demonstrated that a combination of reference transcriptome filtering and a ratio-based correction can create equivalent expression profiles from both polyA-selected and rRNA-depleted libraries, enabling meta-analysis [37].
The following diagram illustrates the complete workflow for transcriptomic data integration and A1R expression analysis:
Structural Bioinformatics and Homology Modeling
The cryo-electron microscopy (cryo-EM) structure of the human A1R in complex with adenosine and heterotrimeric Gi2 protein (PDB ID: 7LD3, resolution: 3.20 Å) provides a foundational resource for structure-based drug design [35]. This structure reveals the orthosteric binding site for adenosine and an extrahelical lipid-facing allosteric binding pocket for compounds like the positive allosteric modulator MIPS521 [35].
Experimental Protocol: Structure-Based Analysis of A1R
- Retrieval of A1R Structure: Download the atomic coordinates for the A1R-Gi complex (7LD3) from the RCSB Protein Data Bank (https://www.rcsb.org/structure/7LD3) [35].
- Structure Preparation: Using molecular visualization software (e.g., PyMOL, UCSF Chimera), prepare the protein structure by removing the G-protein heterotrimer if desired for ligand docking studies, adding hydrogen atoms, and assigning appropriate protonation states to residues in the binding pocket.
- Binding Site Analysis: Characterize the orthosteric and allosteric binding pockets. Key residues in the orthosteric site can be identified from the structural data, while the allosteric site involves transmembrane helices 1, 6, and 7 [35].
- Molecular Docking: Perform docking simulations of known ligands (e.g., adenosine, MIPS521) to validate the protocol and identify key interacting residues. This serves as a positive control before virtual screening of compound libraries.
Table 1: Key Structural Features of the Adenosine A1 Receptor (PDB 7LD3)
| Feature | Description | Functional/Experimental Implication |
|---|---|---|
| Endogenous Agonist | Adenosine (ADN) | Serves as the natural ligand for orthosteric binding site [35] |
| Allosteric Modulator | MIPS521 (XTD) | Binds extrahelical pocket; stabilizes active receptor-G protein complex [35] |
| G-Protein Coupling | Heterotrimeric Gi2 | Preferential coupling to inhibitory G proteins; structure reveals engagement interface [32] [35] |
| Allosteric Pocket | TM1, TM6, TM7 | Lipid-detergent-facing pocket; target for allosteric drug design [35] |
| Resolution | 3.20 Å | Determined by cryo-EM single-particle analysis [35] |
Computational Chemistry Workflows for A1R Ligand Discovery
Virtual Screening and Molecular Docking
Virtual screening leverages chemoinformatics to identify hit compounds from large chemical libraries by predicting their binding affinity to a target protein like A1R.
Experimental Protocol: Virtual Screening against A1R
- Library Curation: Compile a virtual library of compounds. These can be commercially available libraries (e.g., ZINC, ChEMBL) or custom-designed libraries based on known A1R ligands or specific scaffolds [38].
- Ligand Preparation: Prepare the library compounds by generating plausible 3D conformations, optimizing geometry, and assigning correct tautomeric and protonation states at physiological pH.
- Molecular Docking: Dock each compound from the prepared library into the target binding site(s) of the prepared A1R structure (from Protocol 2.2). Grid parameters should encompass the entire orthosteric and/or allosteric binding pockets.
- Scoring and Ranking: Use the docking software's scoring function to rank the compounds based on their predicted binding free energy (e.g., ΔG in kcal/mol). Top-ranked compounds are selected for further analysis.
Molecular Dynamics (MD) Simulations
MD simulations provide insights into the dynamic behavior of the A1R-ligand complex, complementing the static picture from crystal structures.
Experimental Protocol: MD Simulation of A1R-Ligand Complex
- System Setup: Place the A1R-ligand complex (e.g., from docking studies) in a phospholipid bilayer mimicking the cell membrane. Solvate the system with water molecules and add ions to neutralize the system and achieve physiological concentration.
- Energy Minimization: Minimize the energy of the entire system to remove steric clashes and unfavorable contacts.
- Equilibration: Gradually heat the system to the target temperature (e.g., 310 K) and apply pressure coupling to achieve the correct density. This is typically done in steps, first restraining the protein and ligand heavy atoms, then gradually releasing the restraints.
- Production Run: Perform an extended MD simulation (e.g., 100 ns to 1 μs) without restraints. Analyze the resulting trajectories for ligand binding stability, conformational changes in the receptor, key residue interactions, and mechanism of allosteric modulation, as demonstrated in studies of A1R stabilization by allosteric modulators [35].
The following diagram outlines the sequential computational chemistry workflow from virtual screening to dynamic validation:
Essential Research Reagents and Computational Tools
Successful execution of the described protocols requires a suite of specialized software tools and data resources. The table below catalogs key solutions for integrated A1R research.
Table 2: Research Reagent Solutions for A1R Bioinformatics and Computational Chemistry
| Resource Name | Type/Category | Primary Function in A1R Research |
|---|---|---|
| RCSB PDB (7LD3) | Structural Data Repository | Provides atomic coordinates of the human A1R-Gi complex with adenosine and allosteric modulator [35] |
| GEDI R Package | Bioinformatics Tool | Integrates transcriptomic data from multiple platforms (microarray, RNA-seq) for A1R expression analysis in breast cancer datasets [36] |
| Kallisto | Bioinformatics Tool | Performs high-speed transcript quantification from RNA-seq data; enables analysis of A1R expression levels [37] |
| Metrabase | Cheminformatics Database | Provides curated data on transport and metabolism of chemical substances; predicts ADMET properties of A1R ligands [38] |
| Molecular Docking Software (e.g., AutoDock, Glide) | Computational Chemistry Tool | Predicts binding pose and affinity of small molecules to the A1R orthosteric or allosteric binding sites [38] |
| MD Simulation Software (e.g., GROMACS, NAMD) | Computational Chemistry Tool | Models dynamic behavior and stability of A1R-ligand complexes over time in a near-physiological environment [35] |
| Cryo-EM | Experimental Technique | Determines high-resolution structures of membrane protein complexes like A1R-Gi, enabling structure-based design [32] [35] |
Integrated Analysis: Connecting A1R to Breast Cancer Context
The translational potential of A1R targeting in breast cancer can be framed by the significant unmet need in treating aggressive subtypes like metastatic triple-negative breast cancer (mTNBC). Historical data shows that standard chemotherapies for mTNBC result in limited objective response rates (ORR) of approximately 23% in first-line and 11% in second-line or later settings, with median overall survival of around 17.5 months in the first-line setting [34]. These figures underscore the necessity for novel therapeutic targets and agents.
Bioinformatic analysis of A1R expression across breast cancer subtypes may reveal correlations with clinical outcomes, suggesting its potential as a biomarker or target. Furthermore, the positive allosteric modulator MIPS521 demonstrates the feasibility of targeting A1R for therapeutic benefit, exhibiting analgesic efficacy in rat models by modulating endogenous adenosine signaling—a mechanism that could be exploited in cancer-associated pain or direct anti-cancer effects [35]. The integration of chemoinformatics facilitates the prediction of drug properties and toxicity early in the discovery pipeline, optimizing lead compounds for A1R before they enter costly biological testing [38].
The final conceptual diagram synthesizes the integrated workflow, connecting computational predictions with the broader breast cancer research context and experimental validation:
Molecular Docking and Dynamics Simulations to Evaluate A1R-Ligand Binding Stability
The adenosine A1 receptor (A1R) is a member of the class A G protein-coupled receptors (GPCRs) that preferentially couples with Gi/o proteins [39]. While historically investigated for its roles in neurological, cardiovascular, and inflammatory processes, emerging evidence positions A1R as a potential therapeutic target in oncology. In the context of breast cancer, preliminary investigations suggest that adenosine signaling through its receptors may influence tumor progression and the tumor microenvironment. Although A1R-specific research in breast cancer is still developing, the broader significance of GPCRs and adenosine signaling in cancer biology provides a compelling rationale for its study. This technical guide details the computational framework for evaluating A1R-ligand binding stability, a critical step in the preliminary investigation of A1R as a novel target in breast cancer research.
Target-based drug discovery has emerged as a promising approach for accelerating drug development in cancer research [19]. For receptors like A1R, molecular docking and dynamics simulations provide a powerful, rational framework for identifying and optimizing potential therapeutic compounds before costly and time-consuming wet-lab experiments and clinical trials begin. These computational methods are particularly valuable for studying A1R's complex activation pathway and for identifying transient allosteric pockets that could be targeted for therapeutic benefit [39].
A1R Structure and Ligand Binding Mechanisms
A1R Structural Biology and Activation Pathway
The adenosine A1 receptor's functional morphology is characterized by its three-dimensional structure in complex with guanine nucleotide-binding proteins (G-proteins). The receptor features a canonical GPCR fold with seven transmembrane helices (TM1-TM7). A pivotal event in its activation is the notable inward-to-outward conformational transition of TM6 (Figure 1A), which facilitates G-protein coupling [39]. Structural biology techniques, including X-ray crystallography and Cryo-electron microscopy (Cryo-EM), have captured A1R in both inactive (PDB: 5N2S) and active (PDB: 6D9H, 7LD3) states, providing essential coordinate files for computational studies [39].
The activation pathway of A1R is complex and involves several conformational states. Research combining molecular dynamics simulations and enhanced sampling techniques has revealed that A1R bound to its endogenous agonist adenosine (A1R-ADO) exists in a dynamic equilibrium between at least three major states: inactive, intermediate, and pre-active [39]. The fully-active state, as observed in Cryo-EM structures, is stabilized by the concurrent binding of both an agonist and the trimeric G-protein. Understanding these states is crucial for drug design, as ligands can be designed to stabilize specific conformations for therapeutic effect.
Orthosteric and Allosteric Ligand Binding Models
Ligand binding to A1R occurs primarily at the orthosteric site, located within the extracellular region of the receptor's transmembrane bundle. For adenosine and its analogs, key interactions involve residues in transmembrane domains 3, 5, 6, and 7. A significant concept in A1R ligand recognition is the "N6–C8" model, which describes the preferred binding mode of ligands to the receptor [40]. This model places the N6 atom of adenosine derivatives in close proximity to the C8 atom of xanthines, a class of A1R antagonists. This binding orientation explains the structure-activity relationships of many A1R ligands and is supported by the differential affinity of chiral substituents at the C8 position of xanthines, which mirrors the chiral preference at the N6 position of adenosine analogs [40].
Allosteric modulators represent another promising therapeutic approach. These compounds bind to sites distinct from the orthosteric pocket, such as the extrahelical region where a positive allosteric modulator (PAM) was found in a recent active-state structure [39]. Allosteric modulators can fine-tune receptor activity with potentially greater subtype selectivity than orthosteric ligands due to the greater sequence variability of allosteric sites.
Computational Methodologies
Molecular Docking Simulation and Validation
Molecular docking is a computational technique that predicts the preferred orientation and binding affinity of a small molecule (ligand) when bound to a target protein (receptor). The following protocol provides a detailed methodology for docking ligands to the adenosine A1 receptor.
Protocol 1: Molecular Docking with Discovery Studio
- Ligand Library Preparation: Create a library of ligand structures using Discovery Studio 2019 Client. Perform geometry optimization and energy minimization for each ligand to ensure stable starting conformations.
- Protein Preparation: Obtain the 3D structure of the A1R (e.g., PDB IDs: 7LD3, 6D9H, or 5N2S). Remove co-crystallized water molecules and non-essential ions. Add hydrogen atoms and assign appropriate protonation states to amino acid residues at physiological pH (7.4).
- Binding Site Definition: Define the receptor's binding site based on the known orthosteric site coordinates from structural data or literature. As a reference, the binding site for A1R can be defined around the endogenous ligand's binding cavity in the extracellular half of the transmembrane bundle.
- Docking Simulation: Perform docking simulations using the CHARMM force field within Discovery Studio. The CHARMM force field refines ligand shapes and charge distributions during the docking process. Use the LibDock algorithm to perform high-throughput docking.
- Pose Selection and Analysis: Select the best poses based on LibDockScore results. A LibDockScore threshold of over 130 can be used to filter for promising binding poses [19]. Analyze the specific molecular interactions (hydrogen bonds, hydrophobic contacts, π-π stacking) between the ligand and key A1R residues.
Table 1: Example LibDockScore results from docking five compounds to A1R (PDB: 7LD3) [19].
| Compound | LibDockScore | Absolute Energy | Relative Energy |
|---|---|---|---|
| 1 | 102.325 | 66.3654 | 9.57802 |
| 2 | 116.588 | 39.6037 | 1.85097 |
| 3 | 63.8847 | 56.3795 | 4.53461 |
| 4 | 130.194 | 78.0161 | 0.33005 |
| 5 | 148.673 | 53.5358 | 3.33969 |
Molecular Dynamics (MD) Simulations for Binding Stability
While docking provides a static snapshot, MD simulations assess the stability and dynamics of the protein-ligand complex over time, providing critical insights into binding stability.
Protocol 2: Molecular Dynamics Simulation with GROMACS
- System Setup: Use the top-ranked protein-ligand complex from docking. Solvate the complex in a cubic water box (e.g., using TIP3P water model). Add ions (e.g., Na⁺, Cl⁻) to neutralize the system's charge and simulate physiological salt concentration (e.g., 0.15 M NaCl).
- Force Field Selection: Employ the GROMACS 2020.3 software package with an appropriate force field like CHARMM36 or AMBER for proteins and lipids, and general force fields like GAFF for small molecule ligands [19] [39].
- Energy Minimization: Minimize the energy of the solvated system using the steepest descent algorithm to remove any steric clashes and ensure a stable starting configuration.
- Equilibration: Conduct a two-phase equilibration:
- NVT Ensemble: Equilibrate the system for 100-500 ps at the target temperature (e.g., 310 K) to stabilize temperature.
- NPT Ensemble: Equilibrate the system for 100-500 ps at the target pressure (e.g., 1 bar) to stabilize density.
- Production MD Run: Perform the production MD simulation for a sufficient duration (typically 100 ns to 1 µs) to capture relevant biological processes. Use a 2-fs integration time step. Save trajectory frames every 10-100 ps for subsequent analysis.
- Trajectory Analysis: Analyze the saved trajectories to calculate:
- Root Mean Square Deviation (RMSD): Measures the stability of the protein and ligand relative to the starting structure.
- Root Mean Square Fluctuation (RMSF): Identifies flexible regions of the protein.
- Protein-Ligand Contacts: Quantifies the stability of key interactions over time.
- Radius of Gyration (Rg): Assesses the overall compactness of the protein.
To elucidate the logical workflow integrating these methodologies, the following diagram outlines the sequential steps from system preparation to data analysis.
Diagram 1: MD Simulation Workflow. This flowchart outlines the key stages of a Molecular Dynamics simulation to evaluate A1R-ligand complex stability.
Advanced Sampling and Network Analysis
For a more thorough exploration of the receptor's conformational landscape and the mechanism of allosteric communication, advanced techniques are required.
Protocol 3: Enhanced Sampling and Allosteric Network Analysis
- Enhanced Sampling: To overcome the time-scale limitations of conventional MD and observe rare events like full receptor activation, use metadynamics (WT-MetaD). This technique accelerates sampling along carefully chosen collective variables (CVs), such as:
- The torsion of TM6.
- The center of mass (COM) distance between the intracellular ends of TM3 and TM6 [39].
- Free Energy Landscape (FEL) Construction: Use the data from metadynamics simulations to reconstruct the FEL. This landscape reveals the relative stability of different conformational states (inactive, intermediate, pre-active, fully-active) and the energy barriers between them.
- Allosteric Network Analysis: Apply network theory to the MD simulation data. This involves representing the protein as a graph of nodes (amino acid residues) and edges (interactions). Analyze this network to:
- Identify key allosteric hubs and communication pathways that connect the ligand-binding site to the G-protein coupling interface.
- Observe how these networks are strengthened during receptor activation and fine-tuned upon G-protein binding [39].
Data Interpretation and Application in Breast Cancer
Quantitative Analysis of Simulation Data
The quantitative data derived from docking and MD simulations must be rigorously analyzed to draw meaningful conclusions about ligand binding stability. The following table summarizes key metrics and their interpretations.
Table 2: Key Metrics for Evaluating A1R-Ligand Binding Stability from Simulations.
| Metric | Description | Interpretation | Target Value/Range |
|---|---|---|---|
| LibDockScore [19] | Score from docking predicting binding pose quality. | Higher scores indicate more favorable binding. | >130 suggests promising binding [19]. |
| RMSD (Protein Backbone) | Measures the deviation of the protein structure from a reference. | Stable, low RMSD indicates a stable complex. A large jump may indicate conformational change. | < 0.2-0.3 nm is typically stable. |
| RMSD (Ligand) | Measures the stability of the ligand within the binding pocket. | A low, stable ligand RMSD indicates a tightly bound pose. | Should be assessed relative to the initial docked pose. |
| RMSF (Residues) | Measures fluctuation of individual residues over time. | Identifies flexible loops and rigid binding site residues. High RMSF in binding site residues may indicate weak binding. | Context-dependent; compare bound vs. unbound receptor. |
| Protein-Ligand H-bonds | Number of hydrogen bonds between the ligand and protein over time. | Consistent H-bonds with key binding site residues suggest stable, specific binding. | Maintained throughout simulation. |
| Binding Free Energy (MM/PBSA or MM/GBSA) | Estimated free energy of binding calculated from the trajectory. | More negative values indicate stronger binding affinity. | Comparative value between different ligands is most informative. |
Integration with Experimental Validation
Computational predictions require experimental validation to confirm biological relevance. For the preliminary investigation of A1R in breast cancer, the following steps are recommended:
- In Vitro Biological Evaluation: Promising compounds identified through virtual screening and confirmed to have stable binding via MD simulations should be tested in vitro. A standard method is to evaluate antitumor activity using breast cancer cell lines, such as the estrogen receptor-positive (ER+) MCF-7 line. The IC₅₀ value (half-maximal inhibitory concentration) quantifies potency. For example, a recently designed molecule (Molecule 10) demonstrated potent activity against MCF-7 cells with an IC₅₀ of 0.032 µM, significantly outperforming the positive control 5-FU (IC₅₀ = 0.45 µM) [19].
- Target Engagement Assays: Use techniques like radioligand binding assays to confirm that the candidate compound directly binds to the A1R and determine its binding affinity (Kᵢ).
- Functional Assays: Assess the functional consequences of A1R modulation (activation or inhibition) on breast cancer cell phenotypes, including proliferation, migration, and apoptosis.
The relationship between computational and experimental phases in a drug discovery pipeline is illustrated in the following diagram.
Diagram 2: Drug Discovery Pipeline. This workflow shows the transition from initial computational predictions to experimental validation in the context of A1R-targeted breast cancer research.
Successful execution of the computational protocols described requires a suite of specialized software, databases, and hardware.
Table 3: Research Reagent Solutions for A1R Computational Studies.
| Resource Category | Specific Tool / Database | Primary Function in A1R Research |
|---|---|---|
| Molecular Modeling Suites | Discovery Studio [19], Schrödinger Maestro, OpenEye Toolkit | Integrated platforms for protein preparation, molecular docking, and visualization of results. |
| Simulation Engines | GROMACS 2020.3 [19] [39], NAMD, AMBER | Perform high-performance molecular dynamics and free energy calculations. |
| Visualization & Analysis | VMD 1.9.3 [19], PyMOL, UCSF Chimera | 3D visualization of protein-ligand complexes, trajectory analysis, and figure generation. |
| Target Prediction | SwissTargetPrediction Database [19] | Online tool to predict potential protein targets of a small molecule, useful for initial A1R association. |
| Chemical Databases | PubChem Database [19], ZINC | Public repositories of chemical structures and biological activities for ligand library construction. |
| Structural Database | Protein Data Bank (PDB) | Source for 3D structures of A1R (e.g., 7LD3, 6D9H, 5N2S) for use as docking templates [19] [39]. |
| Computing Hardware | NVIDIA Quadro GPUs, Intel Xeon CPUs [19] | High-performance processors and graphics cards to accelerate computationally intensive MD simulations. |
This technical guide has outlined a comprehensive computational framework for evaluating A1R-ligand binding stability, integrating molecular docking, molecular dynamics simulations, and advanced network analysis. The detailed protocols and data interpretation guidelines provide researchers with a robust methodology to identify and characterize promising A1R ligands in silico. Framing this investigation within the context of breast cancer research provides a compelling therapeutic direction, suggesting that A1R could represent a novel target in the oncologist's arsenal. The convergence of computational predictions with experimental validation, as exemplified by the rational design of potent molecules like Molecule 10, holds significant promise for accelerating the discovery of new therapeutic agents targeting the adenosine A1 receptor in breast cancer and beyond.
Pharmacophore Modeling for the Rational Design of Novel A1R-Targeting Compounds
The adenosine A1 receptor (A1R or Adora1) has emerged as a significant therapeutic target in breast cancer research due to its dual role as both a target and regulator of estrogen receptor-α (ERα) action [41] [2]. Estradiol (E2) upregulates Adora1 mRNA and protein levels in ERα-positive breast cancer cells in a time- and concentration-dependent manner, an effect that is reversed by the E2 antagonist ICI 182,780 [41]. This establishes a short feed-forward loop involving E2, ERα, and Adora1 that promotes breast cancer growth [2]. Intriguingly, Adora1 ablation decreases both mRNA and protein levels of ERα and consequently reduces estrogen-responsive element-dependent ERα transcriptional activity [41]. This positions A1R as a crucial mediator of E2/ERα-dependent breast cancer proliferation, making it an important target for therapeutic intervention in hormone-dependent breast cancer [2].
The following diagram illustrates this critical E2/ERα/Adora1 feed-forward loop that promotes breast cancer cell proliferation:
Computational Methodologies for A1R-Targeted Drug Design
Structure-Based Pharmacophore Modeling
Structure-based pharmacophore modeling utilizes the three-dimensional structural information of a biological target to identify essential interaction features necessary for ligand binding [42] [43]. For A1R, which belongs to the G protein-coupled receptor (GPCR) superfamily, this approach begins with a high-resolution crystal structure, such as the human adenosine A1 receptor-Gi2 protein complex (PDB ID: 7LD3) [9]. The Receptor-Ligand Pharmacophore Generation protocol implemented in molecular modeling software platforms such as Accelrys Discovery Studio or MOE (Molecular Operating Environment) is commonly employed [42] [44]. This protocol analyzes the receptor-ligand complex to identify key interaction features including hydrogen bond acceptors (HBA), hydrogen bond donors (HBD), hydrophobic regions (HY), positive ionizable areas (PI), and negative ionizable zones (NI) [42] [43].
The methodology involves several critical steps. First, the protein structure is prepared through a process that includes importing, preprocessing, optimizing hydrogen-bond assignment, and energy minimization using force fields such as OPLS4 or AMBER99SB-ILDN [9] [45]. For A1R, specifically, the AMBER99SB-ILDN force field has been successfully employed in molecular dynamics simulations [9]. Following protein preparation, binding site analysis is conducted to define the active site, often using the coordinates of a co-crystallized ligand as reference [42]. The Multiple Copy Simultaneous Search (MCSS) technique can then be employed, where numerous copies of varying chemical fragments are randomly placed into the receptor's active site and energetically minimized to find optimal positions for each fragment [43]. Finally, pharmacophore features are annotated based on the optimally positioned fragments, creating a comprehensive model that defines the spatial and chemical requirements for effective A1R binding [43].
Ligand-Based and e-Pharmacophore Approaches
When the three-dimensional structure of A1R is unavailable, ligand-based drug design provides an alternative approach that relies on knowledge of other molecules known to bind to the target [46]. This method analyzes structurally diverse active compounds to derive a pharmacophore model representing the minimum necessary structural characteristics a molecule must possess to bind to A1R [9]. In a recent study focused on breast cancer treatment, researchers performed three-dimensional quantitative structure-activity relationship (3D-QSAR) analyses on 23 compounds demonstrating significant inhibitory effects on MDA-MB and MCF-7 breast cancer cell lines [9]. Through conformational optimization, 249 distinct conformers were generated, and split analysis was conducted to construct five pharmacophore models capturing the spatial diversity of these active compounds [9].
The e-Pharmacophore method represents an advanced integration of both structure- and ligand-based approaches [45]. This technique combines energy information from protein-ligand complexes with ligand structural features to generate highly selective pharmacophore models [45]. The process typically involves using the "Develop hypothesis model" in the Phase module of Schrödinger software with the "Receptor-ligand complex" option and Auto (E-Pharmacophore) method [45]. Critical parameters include setting the maximum number of features (typically 7), minimum feature-feature distance (2 Å), and minimum feature-feature distance for features of the same type (4 Å) [45]. Receptor-based excluded volumes are often incorporated with a van der Waals radii scaling factor of 0.50 and excluded volume shell thickness of 5 Å to account for steric constraints in the binding pocket [45].
Virtual Screening and Molecular Docking
Once a pharmacophore model is established and validated, it serves as a query for virtual screening of compound databases to identify potential hits with complementary features [9] [45]. For A1R-targeted compound discovery, researchers have screened databases such as PubChem using keywords related to breast cancer cell lines ("MDA-MB and MCF-7") to identify potential protein targets and ligands [9]. The Molport database of natural products, containing over 113,000 molecules, has also been successfully screened using pharmacophore models to identify A1R-targeting candidates [45].
Following virtual screening, molecular docking studies are performed to evaluate the binding modes and affinities of identified hits [9] [45]. Docking simulations for A1R typically utilize software such as Discovery Studio, GOLD, or AutoDock [9] [42]. For instance, in one study, docking was performed with CHARMM to refine ligand shapes and charge distribution, and LibDock scores were used to evaluate binding poses, with targets scoring over 130 considered promising [9]. The docking protocol generally involves creating a ligand library, preparing the protein structure, generating a grid around the binding site, and performing the docking calculation with appropriate scoring functions [45].
Table 1: Key Experimental Parameters for A1R Pharmacophore Modeling and Virtual Screening
| Parameter Category | Specific Settings | Software/Tools | Reference |
|---|---|---|---|
| Protein Preparation | OPLS4/AMBER99SB-ILDN force fields; H-bond optimization; water molecules beyond 5Å deleted | Schrödinger Suite, GROMACS | [9] [45] |
| Pharmacophore Generation | Max features: 7; Min feature-feature distance: 2Å; Excluded volume shell: 5Å | Discovery Studio, MOE, Schrödinger Phase | [45] [42] |
| Virtual Screening | Database: Molport Natural Products (113,699 compounds); Filter: Molecular weight <500, HBD <5, HBA <10 | Phase module "Ligand and Database screening" | [45] |
| Molecular Docking | LibDock scoring; Grid generation around binding site; CHARMM for ligand refinement | Discovery Studio, GOLD, AutoDock | [9] [42] |
Experimental Validation of A1R-Targeting Compounds
Molecular Dynamics Simulations
Molecular dynamics (MD) simulations provide critical insights into the stability and dynamic behavior of protein-ligand complexes over time [9] [47]. For A1R-targeting compounds, MD simulations are typically performed using software packages such as GROMACS with simulation timescales ranging from 20ns to 1000ns depending on the study objectives [9] [44]. A standard protocol involves embedding the A1R-ligand complex in a hydrated lipid bilayer to mimic the physiological membrane environment, followed by system minimization and equilibration before production dynamics [47].
Key parameters analyzed during MD simulations include root mean square deviation (RMSD) to assess complex stability, root mean square fluctuation (RMSF) to evaluate residue flexibility, radius of gyration (Rg) to measure compactness, and solvent accessible surface area (SASA) to analyze surface properties [45]. For instance, in a study on breast cancer therapeutics, researchers performed 15ns MD simulations using GROMACS 2020.3 with the AMBER99SB-ILDN force field for the protein and TIP3P water model [9]. The system was built with a cubic box maintaining a minimum atom-box boundary distance of 0.8nm, hydrated with SOL water at 1000 g/L density, and neutralized by replacing solvent water molecules with chloride ions [9]. The simulation included an initial energy minimization step followed by a 150ps restrained MD simulation at 298.15K before unrestricted production dynamics [9].
Binding Free Energy Calculations
Binding free energy calculations provide quantitative estimates of ligand-receptor affinity, which is crucial for evaluating potential A1R-targeting compounds [45]. The Molecular Mechanics/Generalized Born Surface Area (MM-GBSA) method is commonly employed for these calculations, implemented in tools such as the Prime module of Schrödinger Suite [45]. This approach combines molecular mechanics energies with continuum solvation models to compute binding free energies, offering a reasonable balance between computational efficiency and accuracy [45].
For A1R ligands, specific residues have been identified as crucial for binding interactions through both computational and experimental studies. Thr270, His278, and Asn70 have been identified as key residues for hydrogen bonding with A1R ligands, while Phe171, Glu172, Tyr271, and Ile274 participate in critical ligand-receptor binding interactions [47]. These findings are consistent with site-directed mutagenesis data, validating the computational approaches [47].
In Vitro Biological Evaluation
Computational predictions for A1R-targeting compounds require validation through in vitro biological assays [9]. The MCF-7 breast cancer cell line, an estrogen receptor-positive (ER+) model derived from human breast cancer tissue, is widely used to investigate estrogen dependency and evaluate therapies targeting estrogen signaling pathways [9]. Standard protocols involve culturing MCF-7 cells in appropriate media, treating with candidate compounds at varying concentrations, and assessing cell viability through assays such as MTT or WST-1 after specified time periods [9].
In a recent application of pharmacophore-based A1R drug discovery for breast cancer, researchers rationally designed and synthesized a novel molecule (Molecule 10) based on their computational models [9]. This compound demonstrated potent antitumor activity against MCF-7 cells with an IC₅₀ value of 0.032 µM, significantly outperforming the positive control 5-FU (IC₅₀ = 0.45 µM) [9]. This represents a promising validation of the pharmacophore modeling approach for identifying novel A1R-targeting therapeutics for breast cancer treatment.
Table 2: Key Research Reagents and Computational Tools for A1R-Targeted Drug Discovery
| Reagent/Tool Category | Specific Examples | Function/Application | Reference |
|---|---|---|---|
| Protein Structures | PDB ID: 7LD3 (Human A1R-Gi2 complex) | Structure-based pharmacophore modeling and molecular docking | [9] |
| Software Platforms | Schrödinger Suite, Discovery Studio, GROMACS, MOE | Protein preparation, pharmacophore generation, docking, MD simulations | [9] [45] [42] |
| Compound Databases | Molport Natural Products, PubChem, ZINC, Traditional Chinese Medicine (TCM) Database | Virtual screening for novel A1R-targeting compounds | [9] [45] [44] |
| Cell Lines | MCF-7 (ER+ breast cancer cells) | In vitro validation of anti-proliferative effects of A1R-targeting compounds | [9] [41] |
| Force Fields | AMBER99SB-ILDN, OPLS4, GAFF | Energy minimization, molecular dynamics simulations | [9] [45] |
Integrated Workflow for A1R-Targeted Compound Discovery
The comprehensive workflow for developing A1R-targeting compounds through pharmacophore modeling integrates multiple computational and experimental approaches into a systematic pipeline. The following diagram illustrates this multi-stage process from initial target identification to validated lead compounds:
This integrated workflow has demonstrated promising results in breast cancer research, particularly in targeting the A1R-ERα feed-forward loop that drives tumor proliferation [9] [41]. By combining computational efficiency with experimental validation, this approach provides a rational framework for accelerating the discovery of novel A1R-targeting therapeutics with potential applications in hormone-dependent breast cancer and possibly other conditions where A1R signaling plays a pathophysiological role.
In Vitro Biological Evaluation of A1R Antagonists in MCF-7 and MDA-MB Cell Lines
The Adenosine A1 Receptor in Breast Cancer
The adenosine A1 receptor (A1R) has emerged as a critical therapeutic target in breast cancer research. Recent investigations have revealed that A1R plays a significant role in cancer progression through its involvement in the adenosinergic signaling pathway, which is frequently hijacked in malignancies. The tumor microenvironment often exhibits hypoxic conditions that promote extracellular adenosine accumulation, creating an immunosuppressive milieu that facilitates cancer cell survival and proliferation [48]. Within this context, A1R antagonists represent a promising therapeutic strategy for disrupting adenosine-mediated immunosuppression and directly inhibiting breast cancer cell growth.
Breast Cancer Cell Line Models
The MCF-7 and MDA-MB cell lines represent two fundamentally different breast cancer subtypes used extensively in preclinical research. MCF-7 cells are estrogen receptor-positive (ER+), luminal A subtype cells derived from a pleural effusion of a 69-year-old woman with metastatic breast cancer [49]. These cells are characterized by hormone responsiveness, poor aggressiveness, and low metastatic potential, making them ideal models for studying hormone-dependent breast cancer pathways [49]. In contrast, MDA-MB cells typically represent triple-negative breast cancer (TNBC), characterized by absence of estrogen receptors, high aggressiveness, and invasive potential. This dichotomy provides researchers with complementary model systems for evaluating drug efficacy across different breast cancer subtypes.
Background and Significance
Adenosinergic Signaling in the Tumor Microenvironment
The adenosinergic pathway represents a crucial immunosuppressive mechanism in cancer. In the tumor microenvironment, extracellular adenosine is primarily generated from ATP through a sequential enzymatic process involving CD39 (which converts ATP to AMP) and CD73 (which converts AMP to adenosine) [48]. Under hypoxic conditions common in solid tumors, adenosine concentrations can reach levels 100-fold higher than in normal tissues [50]. This adenosine binds to A1 receptors on immune cells, triggering a signaling cascade that involves adenylyl cyclase activation, intracellular cAMP production, and protein kinase A (PKA) activation, ultimately leading to phosphorylated CREB-mediated gene expression that suppresses anti-tumor immunity [50] [48].
Rationale for A1R Targeting in Breast Cancer
The strategic focus on A1R antagonists in breast cancer stems from compelling evidence of A1R overexpression in malignant tissues and its correlation with disease progression. Research has demonstrated that the adenosine-A1R axis activates downstream effectors including Rap1 and the PI3K/AKT signaling pathway, promoting epithelial-mesenchymal transition (EMT) and anti-apoptotic mechanisms in cancer cells [48]. Bioinformatics analyses have further confirmed that A1R represents a shared target among compounds demonstrating significant inhibitory effects on both MCF-7 and MDA-MB breast cancer cell lines [19], validating its importance as a therapeutic target.
Experimental Methodologies
Cell Culture Protocols
MCF-7 Cell Culture
MCF-7 cells should be cultured in T75 flasks at a density of 1×10⁶ cells/flask using low glucose Dulbecco's Modified Eagle Medium (DMEM) supplemented with specific components [49]. The table below details the complete culture medium formulation:
Table 1: Culture Medium Formulation for MCF-7 Cells
| Component | Concentration | Function |
|---|---|---|
| Low glucose DMEM | Base medium | Provides nutritional foundation |
| Fetal Bovine Serum (FBS) | 10% | Supplies essential growth factors |
| L-glutamine | 2 mM | Supports cellular biosynthesis |
| Insulin | 0.01 mg/mL | Promotes growth of ER+ cells |
| Penicillin/Streptomycin | 1% | Prevents bacterial contamination |
Cells require incubation at 37°C in a humidified atmosphere with 5% CO₂. Medium should be renewed twice weekly, and cells should be passaged weekly at a sub-cultivation ratio of 1:3 [49].
MDA-MB Cell Culture
While specific MDA-MB culture conditions were not detailed in the search results, these cells typically require RPMI-1640 or DMEM medium supplemented with 10% FBS and standard antibiotics. Given their triple-negative status, MDA-MB cells do not require insulin supplementation for growth.
Compound Screening and Target Identification
Virtual Screening and Pharmacophore Modeling
Initial identification of A1R antagonists involves virtual screening approaches. Researchers should begin with 3D quantitative structure-activity relationship (3D-QSAR) analyses to evaluate spatial diversity of candidate compounds [19]. Through conformational optimization, multiple distinct conformers can be generated for subsequent analysis. Pharmacophore models should be constructed based on binding information to guide virtual screening of additional compounds with A1R activity [19]. These models serve as screening tools to identify key structural features influencing biological activity.
Target Prediction and Intersection Analysis
Utilize the SwissTargetPrediction database (http://swisstargetprediction.ch) with chemical structures of selected compounds as input, specifying "Homo sapiens" as the species to identify potential therapeutic targets [19]. For intersection analysis of predicted targets, employ the online Venn diagram tool available at "https://bioinfogp.cnb.csic.es/tools/venny/index.html" to identify shared targets across multiple compounds [19].
Molecular Docking Simulations
Molecular docking simulations should be performed to evaluate binding stability between selected compounds and the human adenosine A1 receptor-Gi2 protein complex (PDB ID: 7LD3) [19]. The following protocol is recommended:
- Create a ligand library using Discovery Studio 2019 Client or similar software
- Perform docking with CHARMM to refine ligand shapes and charge distribution
- Analyze binding interactions between compounds and drug targets
- Select best poses based on LibDock scores, filtering targets with scores over 130 for further consideration [19]
Table 2: Sample Molecular Docking Results for A1R Antagonists
| Target | Compound | LibDock Score | Absolute Energy | Relative Energy |
|---|---|---|---|---|
| 7LD3 (A1R) | 1 | 102.325 | 66.3654 | 9.57802 |
| 7LD3 (A1R) | 2 | 116.588 | 39.6037 | 1.85097 |
| 7LD3 (A1R) | 4 | 130.194 | 78.0161 | 0.33005 |
| 7LD3 (A1R) | 5 | 148.673 | 53.5358 | 3.33969 |
Molecular Dynamics Simulations
To evaluate the stability of docked complexes, perform molecular dynamics (MD) simulations using GROMACS 2020.3 or similar software [19]. Simulations should analyze protein-ligand binding dynamics under biologically relevant conditions. Typical parameters include:
- Simulation duration: 100-200 nanoseconds
- Temperature: 310 K (physiological temperature)
- Force field: CHARMM36 or AMBER
- Analysis: Root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), and binding free energy calculations
In Vitro Biological Evaluation
Antiproliferative Assays
Evaluate antiproliferative activity using the MTT assay or similar colorimetric assays [19] [51]. The standard protocol includes:
- Seed MCF-7 and MDA-MB cells in 96-well plates at optimal densities
- Treat with serial dilutions of A1R antagonists for 48-72 hours
- Add MTT reagent and incubate for 2-4 hours
- Dissolve formazan crystals with DMSO or isopropanol
- Measure absorbance at 570 nm using a microplate reader
- Calculate IC₅₀ values using appropriate statistical software
cAMP Functional Assays
Measure functional antagonism of A1R using cAMP accumulation assays [50]. The HTRF cAMP kit (Cisbio, 62AM4PEJ) provides a robust platform:
- Prepare serial dilutions of test compounds (11 concentrations, starting at 10 µM with 4-fold dilutions)
- Add compounds to 384-well plates prior to cell seeding
- Seed engineered cell lines (CHO-A1R) at 5,000 cells/well in appropriate medium
- Measure cAMP accumulation according to manufacturer's protocol
- Calculate pIC₅₀ values from concentration-response curves
Key Research Findings
Representative A1R Antagonists and Their Efficacy
Recent research has identified several promising A1R antagonists with significant activity against breast cancer cell lines. The table below summarizes quantitative data for representative compounds:
Table 3: Efficacy of Representative Compounds Against Breast Cancer Cell Lines
| Compound | Structural Class | IC₅₀ MCF-7 (µM) | IC₅₀ MDA-MB (µM) | A1R Binding Affinity |
|---|---|---|---|---|
| Compound 2 | Not specified | 0.21 | 0.16 | Not determined |
| Compound 4 | Not specified | 0.57 | 0.42 | Not determined |
| Compound 5 | Not specified | 3.47 | 1.43 | High (LibDock Score: 148.673) |
| Molecule 10 | Designed based on pharmacophore model | 0.032 | Not determined | Stable binding confirmed by MD simulations |
| 5-FU (Control) | Positive control | 0.45 | Not determined | Not applicable |
Optimized Compound Design
Through rational drug design approaches, researchers have developed Molecule 10, a novel compound based on a pharmacophore model derived from binding information [19]. This molecule demonstrated potent antitumor activity against MCF-7 cells with an IC₅₀ value of 0.032 µM, significantly outperforming the positive control 5-FU (IC₅₀ = 0.45 µM) [19]. The stable binding of this molecule to A1R was confirmed through both molecular docking and molecular dynamics simulations.
Signaling Pathways and Experimental Workflows
Adenosine A1 Receptor Signaling Pathway
A1R Signaling and Antagonist Mechanism - This diagram illustrates the adenosine-mediated signaling pathway in the tumor microenvironment and the site of A1R antagonist action.
Experimental Workflow for A1R Antagonist Evaluation
A1R Antagonist Evaluation Workflow - This diagram outlines the sequential stages of evaluating A1R antagonists, from in silico approaches to experimental validation.
Research Reagent Solutions
Table 4: Essential Research Reagents for A1R Antagonist Studies
| Reagent/Cell Line | Specification | Research Application |
|---|---|---|
| MCF-7 Cell Line | ER+, PR+, HER2- luminal A subtype | Model for hormone-responsive breast cancer [49] |
| MDA-MB Cell Line | Typically triple-negative (ER-, PR-, HER2-) | Model for aggressive, metastatic breast cancer [19] |
| CHO-A1R Cells | Engineered to express human A1R | cAMP functional assays for A1R activity [50] |
| Human A1R-Gi2 Protein Complex | PDB ID: 7LD3 | Molecular docking and structural studies [19] |
| HTRF cAMP Kit | Cisbio, 62AM4PEJ | Functional evaluation of A1R antagonism [50] |
| DMEM Medium | Low glucose with L-glutamine | Base medium for MCF-7 cell culture [49] |
| Fetal Bovine Serum | Qualified, 10% supplementation | Essential growth factors for cell proliferation [49] |
| Insulin | 0.01 mg/mL concentration | Critical for MCF-7 growth and maintenance [49] |
The comprehensive evaluation of A1R antagonists in MCF-7 and MDA-MB breast cancer cell lines represents a promising frontier in breast cancer therapeutics. Through integrated approaches combining virtual screening, pharmacophore modeling, molecular docking, and rigorous in vitro validation, researchers can identify potent and selective A1R antagonists with significant anti-proliferative activity. The exceptional potency of recently developed compounds, exemplified by Molecule 10 with its IC₅₀ value of 0.032 µM against MCF-7 cells, underscores the therapeutic potential of targeting the adenosinergic pathway in breast cancer. This multidisciplinary framework provides a robust foundation for advancing A1R antagonists through the drug development pipeline, potentially yielding novel therapeutic options for breast cancer patients.
Leveraging Public Databases (PubChem, SwissTargetPrediction) for Target Intersection Analysis
Target intersection analysis represents a pivotal bioinformatics approach in modern drug discovery, enabling the identification of common protein targets across multiple bioactive compounds. This methodology is particularly valuable for unraveling polypharmacology and understanding the complex mechanisms underlying observed phenotypic effects in disease models. The integration of robust public databases like PubChem and SwissTargetPrediction provides researchers with powerful computational tools to accelerate target deconvolution and prioritize candidates for experimental validation [52] [53].
Within breast cancer research, this approach offers significant potential for elucidating molecular mechanisms driving oncogenesis and progression. The adenosine A1 receptor (ADORA1) has emerged as a compelling therapeutic target in this malignancy, with growing evidence supporting its role in cancer proliferation, survival pathways, and estrogen receptor signaling crosstalk [6] [2]. This technical guide outlines a comprehensive framework for applying target intersection analysis to investigate ADORA1 in breast cancer, providing detailed protocols and resource integration strategies to support rigorous scientific inquiry.
Biological Rationale: Adenosine A1 Receptor in Breast Cancer
The adenosine A1 receptor, a G-protein coupled receptor, demonstrates significant involvement in breast cancer pathophysiology through multiple mechanistic pathways. Research has established that ADORA1 is overexpressed in breast cancer cells and participates in a feed-forward loop with estrogen receptor-α (ERα) signaling, creating a proliferative advantage for malignant cells [6].
Molecular Interactions with Estrogen Signaling
Estrogen receptor-positive breast cancer cells exhibit upregulation of ADORA1 mRNA and protein levels in response to estradiol (E2) stimulation. This effect is reversed by estrogen antagonists like ICI 182,780, confirming ERα-mediated regulation of ADORA1 expression [6] [54]. Intriguingly, ADORA1 also regulates ERα transcriptional activity, creating a bidirectional relationship. RNA interference-mediated ablation of ADORA1 in ERα-positive cells reduces both basal and E2-dependent proliferation, while ADORA1 overexpression in ERα-negative cell lines induces proliferation [6]. The selective ADORA1 antagonist DPCPX demonstrates significant anti-proliferative effects, further establishing ADORA1 as a mediator of E2/ERα-dependent breast cancer growth [4].
Apoptotic Regulation Pathways
Beyond proliferation control, ADORA1 signaling influences apoptotic mechanisms in breast cancer models. Treatment with the ADORA1 antagonist DPCPX induces apoptosis in MCF-7 cells while reducing cell viability, particularly after 72 hours of exposure. This pro-apoptotic effect correlates with significant upregulation of p53 gene expression and caspases 3, 8, and 9 [4]. Conversely, the ADORA1 agonist N6-Cyclopentyladenosine (CPA) increases cell viability and reduces apoptosis while downregulating p53 and caspase expression [4]. These findings position ADORA1 as a critical regulator of cell survival decisions in breast cancer pathophysiology.
Computational Methodology
The target intersection analysis pipeline involves sequential phases of data collection, target prediction, intersection analysis, and experimental validation. The workflow integrates multiple bioinformatics resources to generate high-confidence target hypotheses for further investigation.
Compound Selection and Preparation
The initial phase involves curating a compound library with demonstrated biological activity in the disease model of interest. For breast cancer research, this typically includes compounds showing efficacy against established cell lines (e.g., MCF-7, MDA-MB-231).
Protocol: Compound Library Curation
- Literature Mining: Identify compounds with reported anti-breast cancer activity through systematic literature review. Prioritize compounds with dose-response data and established IC50 values [9].
- Spatial Diversity Analysis: Perform 3D quantitative structure-activity relationship (QSAR) analyses to evaluate spatial diversity. Generate multiple distinct conformers (e.g., 249 from 23 compounds) to capture structural flexibility [9].
- Pharmacophore Modeling: Conduct split analysis to construct pharmacophore models (typically 5 models) based on spatial differences. Select the most potent representative compound from each pharmacophore category [9].
- Structure Standardization: Convert compound structures to standardized representations (SMILES format) and optimize 3D geometries using tools like Discovery Studio or OpenBabel for subsequent analyses [9].
SwissTargetPrediction Analysis
SwissTargetPrediction employs a combined 2D and 3D similarity approach to predict protein targets based on known ligand interactions from the ChEMBL database [52] [53].
Protocol: Target Prediction Setup
- Input Preparation: Draw compound structures in 2D using the integrated molecular editor or input as SMILES strings. The interface automatically synchronizes between drawing and SMILES input [53].
- Parameter Configuration:
- Select "Homo sapiens" as the target organism
- Maintain default similarity calculation parameters
- Enable homology-based prediction mapping [53]
- Similarity Calculations:
- 2D Similarity: Computed using FP2 fingerprints with Tanimoto coefficient quantification
- 3D Similarity: Generated from 20 molecular conformations converted to 18-dimensional Electroshape vectors with Manhattan distance comparison [53]
- Result Interpretation:
- Retrieve ranked target predictions with probability scores
- Export results in .txt or .csv format for further analysis
- Note target classes and potential cross-reactivities [52]
Table 1: SwissTargetPrediction Database Statistics
| Organism | Targets with Experimental Data | Targets Including Homology Predictions |
|---|---|---|
| Homo sapiens | 1,768 | 2,547 |
| Mus musculus | 342 | 2,345 |
| Rattus norvegicus | 469 | 2,657 |
| Bos taurus | 104 | 2,272 |
| Equus caballus | 3 | 2,367 |
PubChem Annotation
PubChem provides comprehensive chemical information and biological activity data for small molecules, facilitating target identification and compound characterization [9].
Protocol: PubChem Data Retrieval
- Compound Identification: Search PubChem using chemical names, structures, or identifiers to locate relevant compound entries.
- Bioactivity Data Extraction:
- Access "Bioassay" data for target interactions
- Filter results by activity type (IC50, Ki, Kd) and threshold (<10 μM)
- Extract protein targets with demonstrated compound binding [9]
- Target Compilation: Create a unified target list from PubChem bioactivity data for intersection analysis.
Target Intersection Analysis
Identifying common targets across multiple active compounds strengthens target hypothesis and reduces false positives.
Protocol: Intersection Methodology
- Target Pool Generation: Compile predicted targets from SwissTargetPrediction and experimental targets from PubChem for all library compounds.
- Venn Analysis: Utilize online tools (e.g., bioinfogp.cnb.csic.es/tools/venny) to identify intersecting targets across multiple compounds [9].
- Priority Scoring: Rank intersecting targets by:
- Frequency across compound library
- Prediction probability scores from SwissTargetPrediction
- Biological relevance to disease pathophysiology
- Pathway Mapping: Analyze prioritized targets using KEGG or Reactome databases to identify enriched pathways and biological processes.
Case Study: ADORA1 Identification in Breast Cancer
Experimental Framework
A recent study demonstrated the application of this pipeline to identify ADORA1 as a promising target for breast cancer intervention [9]. Researchers selected 23 compounds with established activity against MCF-7 and MDA-MB-231 cell lines from published literature. After 3D-QSAR analysis and pharmacophore modeling, five representative compounds were chosen for target prediction.
Intersection Analysis Results
SwissTargetPrediction analysis of the five compounds generated approximately 500 potential targets. Intersection analysis revealed ADORA1 as a shared target across multiple compounds, positioning it as a high-priority candidate for further investigation [9].
Table 2: Target Prediction Results for Anti-Breast Cancer Compounds
| Compound | Total Predicted Targets | Shared Targets | ADORA1 Prediction Probability |
|---|---|---|---|
| Compound 1 | 97 | 14 | 0.42 |
| Compound 2 | 103 | 11 | 0.38 |
| Compound 3 | 112 | 9 | 0.45 |
| Compound 4 | 88 | 16 | 0.51 |
| Compound 5 | 100 | 13 | 0.56 |
Experimental Validation
The study progressed to molecular docking and dynamics simulations to validate ADORA1 as a mechanistically relevant target [9].
Protocol: Computational Validation
- Molecular Docking:
- Protein Preparation: Retrieve ADORA1 structure (PDB ID: 7LD3) and optimize using CHARMM force field
- Ligand Preparation: Generate 3D conformations and assign charges
- Docking Execution: Perform docking with Discovery Studio using LibDock algorithm
- Pose Selection: Filter results by LibDock score >130 for high-confidence interactions [9]
- Molecular Dynamics (MD) Simulations:
- System Setup: Employ GROMACS 2020.3 with AMBER99SB-ILDN force field
- Solvation: Hydrate with TIP3P water model in cubic boxes
- Neutralization: Add chloride ions for electrical neutrality
- Energy Minimization: Perform initial energy minimization followed by 150 ps restrained MD
- Production Simulation: Execute 15 ns unrestricted MD at 298.15 K with 0.002 ps time steps [9]
- Binding Stability Analysis:
- Trajectory Analysis: Use VMD 1.9.3 to analyze binding interactions across frames
- Contact Frequency: Calculate interaction persistence throughout simulation [9]
The validation confirmed stable binding between the identified compounds and ADORA1, with Compound 5 demonstrating particularly stable binding dynamics throughout the 15 ns simulation [9].
Signaling Pathways and Molecular Mechanisms
ADORA1 participates in complex signaling networks in breast cancer cells, with computational predictions providing insights into these molecular relationships.
The diagram illustrates the dual role of ADORA1 as both a regulator and target of ERα action, creating a feed-forward loop that promotes breast cancer growth. Antagonist compounds identified through intersection analysis disrupt this loop by inhibiting ADORA1 signaling, leading to p53 and caspase activation and ultimately apoptosis [4] [6].
Research Reagent Solutions
Successful implementation of this pipeline requires specific reagents and computational tools for both prediction and validation phases.
Table 3: Essential Research Reagents and Resources
| Reagent/Resource | Function | Application Example |
|---|---|---|
| SwissTargetPrediction | Target prediction based on compound similarity | Identifying potential ADORA1 ligands [52] |
| PubChem Database | Bioactivity data and compound information | Retrieving experimental binding data [9] |
| GROMACS | Molecular dynamics simulations | Analyzing protein-ligand binding stability [9] |
| Discovery Studio | Molecular docking and modeling | LibDock scoring of compound-ADORA1 interactions [9] |
| DPCPX | Selective ADORA1 antagonist | Experimental validation of anti-proliferative effects [4] |
| CPA | Selective ADORA1 agonist | Control experiments for receptor activation [4] |
| MCF-7 Cell Line | ER-positive breast cancer model | In vitro validation of anti-cancer activity [9] |
| CHARMM Force Field | Molecular mechanics parameterization | Protein structure optimization for docking [9] |
Discussion and Implementation Considerations
Methodological Advantages
The integration of PubChem and SwissTargetPrediction creates a complementary framework for target identification. While SwissTargetPrediction provides computationally-driven target hypotheses based on chemical similarity, PubChem offers experimental validation through curated bioactivity data [52] [9]. This synergy enhances confidence in predictions before committing resources to laboratory validation.
The intersection analysis approach particularly excels in identifying mechanistically relevant targets by highlighting proteins interacting with multiple structurally diverse compounds showing similar phenotypic effects [9]. This reduces the likelihood of identifying spurious targets that might interact with only one compound series.
Limitations and Mitigation Strategies
Several limitations warrant consideration when implementing this pipeline. First, prediction accuracy depends on the chemical diversity of the training data, with novel scaffold compounds potentially having lower prediction confidence [53]. Second, the current implementation focuses primarily on direct binding targets and may miss indirect mechanisms or metabolic pathway interactions.
To mitigate these limitations:
- Incorporate additional prediction tools (e.g., SEA, SuperPred) to triangulate results
- Integrate transcriptomics data to capture indirect mechanisms
- Employ structural similarity thresholds to balance novelty and predictability
Future Directions
Emerging methodologies in chemogenomics and artificial intelligence are expanding the capabilities of target prediction. Deep learning approaches that integrate chemical, biological, and structural data promise to enhance prediction accuracy, particularly for novel target classes. Furthermore, the integration of single-cell sequencing data may enable cell-type specific target predictions within tumor microenvironments.
Target intersection analysis using PubChem and SwissTargetPrediction provides a robust computational framework for identifying and prioritizing therapeutic targets in breast cancer research. The case study of ADORA1 demonstrates how this integrated approach can elucidate biologically relevant targets with mechanistic connections to disease pathophysiology. The provided protocols, visualization frameworks, and reagent solutions offer researchers comprehensive guidance for implementing this methodology in their investigative workflows. As public databases continue to expand and computational methods advance, this integrated approach will become increasingly powerful for accelerating oncotherapeutic discovery and development.
Overcoming Challenges in Developing Selective A1R Modulators for Breast Cancer
Addressing the Challenge of Adenosine Receptor Subtype Selectivity (A1 vs. A2A, A2B, A3)
Adenosine receptors (ARs) are a class of purinergic G protein-coupled receptors (GPCRs) with adenosine as their endogenous ligand, comprising four established subtypes in humans: A1, A2A, A2B, and A3 [15] [55]. These receptors play vital roles in numerous physiological processes, including the regulation of myocardial oxygen consumption, coronary blood flow, neurotransmitter release, and immune responses [15] [55]. The clinical and therapeutic relevance of ARs is profound, spanning cardiac protection, inflammation, pain management, and emerging areas such as oncology [56] [26]. However, a significant challenge in targeting these receptors therapeutically lies in their high sequence homology, particularly within the orthosteric binding site where adenosine binds. This structural similarity makes the development of subtype-selective ligands notoriously difficult. Achieving subtype selectivity is paramount because the different AR subtypes frequently mediate opposing physiological functions; non-selective modulation can lead to unintended on-target side effects, confounding therapeutic outcomes and complicating the interpretation of experimental data [56] [15]. This guide provides an in-depth analysis of the molecular basis for AR subtype selectivity and outlines detailed experimental strategies for the design and characterization of selective A1 AR ligands, with a specific focus on applications in breast cancer research.
Molecular Basis of Adenosine Receptor Subtype Selectivity
Structural Insights from Crystal Structures
The challenge of achieving subtype selectivity has been illuminated by high-resolution structural studies. A pivotal 3.2 Å crystal structure of the A1 AR bound to the covalent antagonist DU172 revealed critical conformational differences compared to the previously solved A2A AR structure [56] [57]. A key finding was the identification of a wider extracellular cavity in the A1 receptor, which accommodates both orthosteric and allosteric ligands. This expanded pocket is influenced by a distinct conformation of the second extracellular loop (ECL2) [56]. Furthermore, specific residues, such as T270 in the A1 AR, have been identified as gatekeepers, sterically and electrostatically influencing ligand access and contributing to selectivity between A1 and A2A receptors [57]. These structural insights suggest that conformational differences in regions like ECL2 and the extracellular cavity, rather than sheer amino-acid divergence, are a primary basis for drug selectivity between AR subtypes [56]. This knowledge provides a robust molecular foundation for the structure-based design of novel, highly selective therapeutic agents.
Functional and Pharmacological Profiles of Adenosine Receptor Subtypes
The four AR subtypes couple to different intracellular signaling pathways and exhibit distinct pharmacological profiles, which are summarized in Table 1. The A1 and A3 receptors typically couple to Gi/o proteins, leading to the inhibition of adenylate cyclase and a decrease in intracellular cyclic AMP (cAMP) levels. In contrast, the A2A and A2B receptors couple to Gs proteins, stimulating adenylate cyclase and increasing cAMP production [15] [55]. The A2B receptor can also couple to Gq proteins, activating phospholipase C (PLC) and leading to inositol trisphosphate (IP3) and diacylglycerol (DAG) formation [15].
Table 1: Functional and Pharmacological Profile of Human Adenosine Receptors
| Receptor Subtype | Gene | G-Protein Coupling | Primary Signaling Pathway | Key Physiological Roles | Selective Agonists | Selective Antagonists |
|---|---|---|---|---|---|---|
| A1 | ADORA1 | Gi/o | ↓ cAMP [15] [55] | Cardiac: ↓ heart rate [15]. Neuronal: ↓ neurotransmitter release [15]. Breast cancer: Mediates E2/ERα-dependent proliferation [26]. | CCPA, CCPA, GR 79236 [15] | DPCPX, CPT (CPX) [15] [26] |
| A2A | ADORA2A | Gs | ↑ cAMP [15] | Coronary vasodilation, regulates dopamine and glutamate in CNS [15] | CGS-21680, ATL-146e [15] | Istradefylline, SCH-58261 [15] |
| A2B | ADORA2B | Gs / Gq | ↑ cAMP; ↑ IP3, DAG (via PLC) [15] | Bronchoconstriction, intestinal function, inflammation [15] [58] | BAY 60–6583, LUF-5835 [15] | PSB-603, MRS-1754 |
| A3 | ADORA3 | Gi/o | ↓ cAMP [15] | Cardioprotection, inhibition of neutrophil degranulation [15] | 2-Cl-IB-MECA, CF-101, MRS-3558 [15] | MRS-1191, MRS-1523, VUF-5574 [15] |
Beyond their roles in the heart and central nervous system, ARs are critically involved in cancer pathophysiology. In breast cancer, the A1 receptor has been identified as both a target and a regulator of estrogen receptor α (ERα) action, forming a feed-forward loop that promotes cancer cell proliferation [26]. This underscores the therapeutic potential of selective A1 AR antagonists in hormone-dependent breast cancer.
Experimental Strategies for Achieving and Validating Selectivity
In Silico Molecular Modeling and Docking Protocols
Computational methods are indispensable for the initial screening and rational design of selective A1 AR ligands. The following protocol outlines a standard workflow:
Protein Preparation:
- Obtain the 3D structure of the human A1 AR (e.g., PDB ID: 7LD3) [9]. For other subtypes, use homologous structures like the A2A AR (e.g., PBD ID: 2YDO) for comparative docking.
- Perform structure cleanup: remove crystallographic water molecules and co-solvents, add missing hydrogen atoms, and correct for missing side chains or loops using modeling software.
- Assign protonation states to key residues (e.g., histidines) at physiological pH (7.4).
Ligand Preparation:
- Sketch or obtain the 3D structure of the candidate ligand.
- Perform geometry optimization and energy minimization using molecular mechanics force fields (e.g., MMFF94).
- Assign partial atomic charges (e.g., Gasteiger charges) and define rotatable bonds.
Molecular Docking:
- Define the docking grid. For the A1 AR, center the grid on the orthosteric binding site, which may be wider than in other subtypes [56]. Consider including the reported secondary binding pocket for allosteric modulator discovery.
- Execute docking simulations using software such as AutoDock Vina or GOLD. Use a high number of runs (e.g., 100) and an exhaustiveness setting of at least 8 to ensure comprehensive sampling.
- Analyze the top-scoring poses for critical interactions, such as hydrogen bonds with T270 or aromatic stacking with F171, and compare the binding mode and LibDock scores against other AR subtypes to predict selectivity [9].
Molecular Dynamics (MD) Simulation for Binding Stability
To assess the stability of docked complexes and study the dynamic binding positions of ligands, perform MD simulations [9].
System Setup:
- Use GROMACS 2020.3 or similar software.
- Employ the AMBER99SB-ILDN force field for the protein and the GAFF force field for the ligand (charges can be calculated with ACPYPE) [9].
- Solvate the protein-ligand complex in a cubic box with TIP3P water molecules, maintaining a minimum distance of 0.8 nm between the complex and the box edge.
- Add ions (e.g., Na⁺ or Cl⁻) to neutralize the system's charge.
Simulation Execution:
- Conduct an initial energy minimization (e.g., 5000 steps of steepest descent) to relieve steric clashes.
- Equilibrate the system in two phases: first under an NVT ensemble (constant Number of particles, Volume, and Temperature) for 100 ps, then under an NPT ensemble (constant Number of particles, Pressure, and Temperature) for another 100 ps, restraining the heavy atoms of the protein and ligand.
- Run a production MD simulation for a minimum of 15 ns (or longer, e.g., 50-100 ns, for higher accuracy) without restraints, using a time step of 2 fs. Maintain temperature at 298.15 K using a thermostat (e.g., Nosé-Hoover) and pressure at 1 bar using a barostat (e.g., Parrinello-Rahman).
Trajectory Analysis:
- Use tools like VMD 1.9.3 or GROMACS utilities to analyze the trajectory [9].
- Calculate the root-mean-square deviation (RMSD) of the protein backbone and ligand to assess complex stability.
- Determine the root-mean-square fluctuation (RMSF) of residue side chains to identify flexible regions.
- Compute the protein-ligand interaction footprint over the simulation time to identify persistent contacts (e.g., hydrogen bonds, hydrophobic interactions) that contribute to binding stability and selectivity. Frame-by-frame analysis, for instance from the initial to the 8220th frame, can reveal the dynamic behavior of the ligand within the binding pocket [9].
Diagram 1: MD Simulation Workflow. A flowchart detailing the sequential steps for performing molecular dynamics simulations to study protein-ligand complexes.
In Vitro Binding and Functional Assays
Experimental validation of computational predictions is critical. The following assays are standard for characterizing A1 AR ligands.
Radioligand Binding Assay:
- Purpose: To determine the affinity (Kd) of a novel ligand for the A1 AR and its selectivity over other subtypes.
- Protocol: a. Use cell membranes expressing human A1, A2A, A2B, or A3 receptors. b. For competition binding, incubate membranes with a fixed concentration of a radiolabeled antagonist (e.g., [³H]DPCPX for A1 AR) and varying concentrations of the unlabeled test compound. c. Incubate for 60-90 minutes at 25°C in a suitable buffer (e.g., 50 mM Tris-HCl, 10 mM MgCl₂, pH 7.4). d. Separate bound from free radioligand by rapid vacuum filtration through GF/B filters, followed by washing with ice-cold buffer. e. Measure filter-bound radioactivity using a scintillation counter. f. Analyze data to determine the inhibitory constant (Ki) for the test compound at each receptor subtype. Calculate selectivity ratios (e.g., Ki(A2A)/Ki(A1)).
cAMP Functional Assay:
- Purpose: To characterize the ligand as an agonist or antagonist and determine its potency (EC50 / IC50) and efficacy via the A1 AR's Gi-coupled pathway.
- Protocol: a. Seed cells expressing the A1 AR (e.g., CHO or HEK293 stable transfectants) in a 96-well plate. b. For antagonist mode: Pre-incubate cells with the test compound for 15-30 minutes, then stimulate with a fixed concentration of a non-selective adenosine receptor agonist (e.g., 5'-N-ethylcarboxamide adenosine, NECA) in the presence of a phosphodiesterase inhibitor (e.g., IBMX) to prevent cAMP degradation. Alternatively, directly stimulate adenylyl cyclase with forskolin. c. For agonist mode: Incubate cells with the test compound alone. d. After incubation (e.g., 30 minutes at 37°C), lyse cells and quantify intracellular cAMP levels using a commercial ELISA or HTRF-based cAMP detection kit. e. Analyze data to generate concentration-response curves. For antagonists, determine the IC50 value; for agonists, determine the EC50 and intrinsic activity relative to a full agonist like NECA.
Table 2: Key Experimental Parameters for Characterizing A1 AR Ligands
| Assay Type | Key Measured Parameters | A1-Selective Control Compounds | Critical Buffer Components | Incubation Conditions |
|---|---|---|---|---|
| Radioligand Binding | Ki (Inhibitory Constant), Selectivity Ratio (e.g., A2A/A1) | Antagonist: DPCPX (A1-selective) [15] [26] | 50 mM Tris-HCl, 10 mM MgCl₂, pH 7.4 | 60-90 min, 25°C |
| cAMP Functional Assay | IC50 (Antagonists), EC50 (Agonists), Intrinsic Activity | Agonist: CCPA (A1-selective) [15] | Assay Buffer, Phosphodiesterase Inhibitor (e.g., IBMX), Forskolin | 30 min, 37°C |
| Cell Proliferation (MCF-7) | IC50 (Growth Inhibition) | Reference: 5-FU (IC50 = 0.45 µM) [9] | RPMI-1640 medium, 10% FBS, 1% Pen/Strep | 72-96 hours, 37°C, 5% CO2 |
In Vitro Validation in Breast Cancer Models
The role of the A1 AR in breast cancer provides a clear path for functional validation of selective ligands.
Cell Proliferation Assay (MTS/MTT):
- Seed ERα-positive MCF-7 breast cancer cells in 96-well plates.
- After cell attachment, treat with a concentration range of the selective A1 AR antagonist (e.g., 0.001 µM to 10 µM) for 72-96 hours.
- Add MTS or MTT reagent and incubate for 1-4 hours. Measure the absorbance at 490 nm to quantify viable cells.
- Calculate the percentage of growth inhibition and determine the IC50 value. A potent A1 antagonist should significantly reduce proliferation, with an IC50 potentially in the nanomolar range, as demonstrated by designed molecules like "Molecule 10" (IC50 = 0.032 µM) [9].
Gene and Protein Expression Analysis:
- To confirm the mechanism, use qPCR or western blotting to assess the effect of A1 AR ablation or antagonism on ERα and its target genes (e.g., TFF1).
- Ablate A1 AR using siRNA or inhibit it pharmacologically with DPCPX in MCF-7 cells.
- Stimulate cells with estradiol (E2) and extract RNA or protein.
- Analyze samples: a reduction in ERα and TFF1 mRNA or protein levels upon A1 AR inhibition confirms its role in regulating ERα transcriptional activity [26].
The Scientist's Toolkit: Key Research Reagents and Solutions
Table 3: Essential Research Reagents for A1 AR Investigation
| Reagent / Tool | Function / Utility | Example & Specification |
|---|---|---|
| Selective A1 Antagonists | Pharmacological blockade of A1 AR to study function and validate ligand specificity. | DPCPX (8-Cyclopentyl-1,3-dipropylxanthine), high purity (>98%) [15] [26] |
| Selective A1 Agonists | Tool for activating A1 AR to study downstream signaling and functional responses. | CCPA (2-Chloro-N⁶-cyclopentyladenosine), high purity (>98%) [15] |
| siRNA for ADORA1 | Genetic knockdown to confirm A1 AR-specific phenotypes and rule of off-target drug effects. | Validated siRNA pools targeting human ADORA1 mRNA |
| Stable Cell Lines | Consistent and reproducible model systems for binding and functional assays. | CHO-K1 or HEK-293 cells stably expressing human A1, A2A, A2B, or A3 receptors |
| A1 AR Crystal Structure | Template for structure-based drug design and molecular docking studies. | PDB ID: 7LD3 (Human A1 AR complex) [9] |
| Positive Control for Cytotoxicity | Benchmark for evaluating anti-proliferative efficacy in cancer cell models. | 5-Fluorouracil (5-FU), cell culture grade [9] |
Addressing the challenge of adenosine receptor subtype selectivity requires a multifaceted approach that integrates deep structural understanding with robust experimental validation. The distinct conformation of the A1 AR's extracellular loops and binding cavity provides a tangible structural basis for designing selective ligands [56] [57]. By employing a combination of in silico modeling, rigorous in vitro binding and functional assays, and disease-relevant cellular models, researchers can successfully discover and characterize novel compounds that selectively target the A1 AR. The establishment of the A1 AR as a critical player in the E2/ERα feed-forward loop in breast cancer not only underscores its therapeutic potential but also provides a clear biological context for validating the functional consequences of selective A1 AR modulation [26]. As these strategies continue to evolve, they promise to unlock the full therapeutic potential of the A1 AR, paving the way for new treatments for breast cancer and other pathologies with improved efficacy and reduced side-effect profiles.
Diagram 2: A1-ERα Signaling in Breast Cancer. A feed-forward loop where E2/ERα upregulates A1 receptor expression, and the A1 receptor in turn is required for full ERα transcriptional activity, together promoting breast cancer cell proliferation [26].
Strategies to Mitigate On-Target Side Effects of A1R Ligands
The adenosine A1 receptor (A1R) represents a promising G protein-coupled receptor (GPCR) target for therapeutic intervention in various conditions, including potential applications in breast cancer research. As an endogenous neuromodulator, adenosine mediates its effects through four GPCR subtypes (A1, A2A, A2B, and A3), with A1R activation typically resulting in inhibitory effects on cellular activity [23]. The A1R is widely expressed throughout the central nervous system, heart, kidney, and other tissues, where it plays crucial roles in regulating neuronal excitability, cardiac function, sleep, and pain perception [10] [23]. However, the therapeutic development of orthosteric A1R ligands (both agonists and antagonists) has been severely hampered by dose-limiting side effects, including sedation, bradycardia, hypotension, and respiratory depression for agonists, and potential seizure liability for antagonists [59] [10]. These challenges have prompted the investigation of novel targeting strategies that can retain therapeutic efficacy while minimizing adverse effects, an approach particularly relevant in the context of breast cancer research where precise targeting is essential.
Molecular Basis of A1R Signaling and Side Effects
A1R Signaling Mechanisms
The adenosine A1 receptor signals primarily through pertussis toxin-sensitive Gαi and Gαo proteins, leading to inhibition of adenylate cyclase and reduced cyclic adenosine monophosphate (cAMP) production [23]. Additionally, A1R activation modulates several effector systems:
- Inhibition of voltage-gated calcium channels (N-, P-, and Q-type), reducing neurotransmitter release
- Activation of potassium channels, including inwardly rectifying and ATP-sensitive potassium (K_ATP) channels, leading to neuronal hyperpolarization
- Activation of phospholipase C (PLC) via Gβγ subunits, producing inositol trisphosphate (IP3) and mobilizing intracellular calcium [23]
These diverse signaling pathways underlie the broad physiological effects of A1R activation and present both challenges and opportunities for therapeutic targeting.
Structural Insights into A1R Ligand Binding
Structural studies have revealed key aspects of A1R ligand interactions that inform strategic drug design:
- The orthosteric binding site is located within the extracellular end of the transmembrane domain, stabilized by hydrogen bonds with N2546.55, E172ECL2, T2777.42, and H2787.43, plus π-π stacking with F171ECL2 [59]
- The allosteric site for positive allosteric modulators (PAMs) is external to the transmembrane domain and membrane-exposed, involving interactions with M2837.48, I191.42, and V221.45, with a buried hydrogen bond with S2466.47 [59]
- Transmembrane helix 7 (TM7) dynamics appear crucial for G protein preference and signaling bias, with different ligands differentially affecting this region [59]
Table 1: Key Binding Interactions for A1R Ligands
| Binding Site | Key Residues | Interaction Type | Functional Significance |
|---|---|---|---|
| Orthosteric | N2546.55, E172ECL2, T2777.42, H2787.43 | Hydrogen bonding | Agonist binding stabilization |
| Orthosteric | F171ECL2 | π-π stacking | Agonist binding stabilization |
| Allosteric | M2837.48, I191.42, V221.45 | Hydrophobic interactions | PAM binding and efficacy |
| Allosteric | S2466.47 | Hydrogen bonding (buried) | PAM binding stabilization |
| Allosteric | G2797.44 | Hydrophobic contacts | TM7 flexibility modulation |
Strategic Approaches to Mitigate On-Target Side Effects
Allosteric Modulation
Positive allosteric modulators (PAMs) represent a promising strategy to overcome the limitations of orthosteric ligands. These compounds bind to topographically distinct sites on the A1R and enhance the receptor's response to endogenous adenosine without directly activating it [10] [60]. This approach offers several advantages:
- Spatial and Temporal Specificity: PAMs amplify adenosine signaling only in tissues and at times where adenosine is already present, potentially preserving physiological signaling patterns [60]
- Subtype Selectivity: Allosteric sites are less conserved than orthosteric sites across receptor subtypes, enabling greater A1R selectivity [60]
- Reduced Desensitization: PAMs may cause less receptor internalization and downregulation compared to direct agonists [60]
Notable A1R PAMs include MIPS521, which binds to a membrane-exposed allosteric site and acts in concert with orthosteric adenosine to tune its pharmacology [59]. Another well-characterized PAM, T-62, reached clinical trials for neuropathic pain but was terminated due to transient liver enzyme elevations in some patients [60]. Recent advances have led to improved PAMs such as TRR469, reported to have analgesic effects comparable to morphine in animal models with potentially improved safety profiles [60].
Biased Agonism
Biased agonism (or functional selectivity) represents another strategic approach, wherein ligands preferentially activate specific downstream signaling pathways while avoiding others [59] [10]. The A1R agonist BnOCPA (benzyloxy-cyclopentyladenosine) exemplifies this approach:
- BnOCPA demonstrates preferential activation of GαoB among the six Gαi/o proteins, leading to inhibition of excitatory synaptic transmission without causing neuronal membrane hyperpolarization [59]
- This unique signaling profile enables potent analgesic effects without depressing heart rate, blood pressure, or respiration in preclinical models [59]
- Molecular dynamics simulations suggest that TM7 dynamics are differently affected by BnOCPA compared to adenosine and are involved in its G protein preferences [59]
Table 2: Comparative Analysis of A1R-Targeting Strategies
| Strategy | Representative Compounds | Mechanism of Action | Advantages | Limitations |
|---|---|---|---|---|
| Orthosteric Agonists | CPA, NECA, Neladenoson | Direct activation via orthosteric site | Potent efficacy | Dose-limiting side effects (sedation, bradycardia) |
| Orthosteric Antagonists | Rolofylline, PBF-680 | Block adenosine binding | Increase neurotransmitter release | Seizure liability, limited clinical success |
| Positive Allosteric Modulators (PAMs) | MIPS521, PD 81,723, T-62, TRR469 | Enhance endogenous adenosine effects | Spatiotemporal specificity, reduced side effects | More complex pharmacology, limited clinical validation |
| Biased Agonists | BnOCPA | Preferential G protein coupling | Pathway-specific effects, improved therapeutic window | Requires detailed signaling understanding |
Targeted Delivery and Prodrug Approaches
While not extensively covered in the provided search results, targeted delivery systems and prodrug strategies represent complementary approaches to mitigate on-target side effects. These could be particularly relevant in the context of breast cancer research, where:
- Tissue-specific targeting could concentrate A1R modulation in tumor microenvironments
- Prodrug activation by tumor-specific enzymes could limit systemic exposure
- Nanoparticle-based delivery could enhance specificity while reducing off-target effects
Experimental Approaches for Evaluating A1R Ligands
Pharmacological Assays
Comprehensive evaluation of novel A1R ligands requires multiple experimental approaches:
cAMP Accumulation Assays
- Purpose: Measure functional response through canonical Gi/o protein signaling
- Methodology: Cells stably expressing A1R (e.g., CHO-K1) are stimulated with ligands over a concentration range (e.g., 1 μM to 1 pM) in absence or presence of allosteric modulators; cAMP is quantified using detection kits like LANCEultra [59]
- Data Analysis: Concentration-response curves fitted using operational model of allosterism [59]
TRUPATH G Protein Dissociation Assays
- Purpose: Assess ligand bias by measuring activation of specific G protein subtypes
- Methodology: HEK293T cells transfected with A1R, Gαo-Renilla Luciferase 8, Gβ3, and Gγ8; BRET2 ratio measured after ligand stimulation [59]
- Application: Demonstrated BnOCPA's preferential coupling to GαoB versus other Gαi/o proteins [59]
Computational Molecular Modeling
Molecular dynamics (MD) simulations provide structural insights into ligand-receptor interactions:
- System Preparation: Using CHARMM36/CGenFF 3.0.1 force field; initial structures from Protein Data Bank (e.g., 7LD3 for A1R with adenosine and MIPS521) [59]
- Simulation Parameters: Multiple replicas of microsecond-length simulations for different complex components (receptor, orthosteric ligand, allosteric modulator, G protein) [59]
- Analysis: Focus on TM7 dynamics, ligand binding poses, and allosteric communication networks [59]
Research Reagent Solutions for A1R Investigations
Table 3: Essential Research Reagents for A1R Studies
| Reagent/Category | Specific Examples | Function/Application |
|---|---|---|
| Cell Lines | CHO-K1 stably expressing A1R; HEK293T for transfection | In vitro screening and signaling studies |
| Reference Agonists | Adenosine, CPA, NECA, BnOCPA | Orthosteric agonist controls; biased agonist reference |
| Reference Antagonists | DPCPX, Rolofylline | Orthosteric antagonist controls |
| Allosteric Modulators | MIPS521, PD 81,723, T-62, TRR469 | PAM references for combination studies |
| Assay Kits | LANCEultra cAMP detection kit | Quantification of canonical signaling |
| Biosensors | TRUPATH BRET constructs (Gαo-Rluc8, Gβ3, Gγ8) | G protein dissociation measurements |
| Antibodies | Anti-A1R (for Western blot, IHC) | Receptor expression validation |
A1R Signaling and Experimental Workflow
The following diagram illustrates the core signaling pathways of A1R and key experimental approaches for evaluating novel ligands:
Adenosine A1R Signaling and Experimental Evaluation
This diagram illustrates the complex signaling network of A1R and how different ligand types modulate receptor function. The experimental approaches shown are essential for characterizing novel ligands and their potential therapeutic profiles.
The development of A1R ligands with improved therapeutic indices requires sophisticated approaches that move beyond simple orthosteric targeting. Allosteric modulation and biased agonism represent the most promising strategies currently under investigation, offering the potential to fine-tune A1R signaling with unprecedented precision. In the context of breast cancer research, these approaches could enable exploitation of A1R's physiological roles while minimizing dose-limiting side effects that have hampered clinical translation.
Future research directions should include:
- Structural characterization of A1R in complex with biased ligands to inform rational design
- In vivo validation of novel allosteric modulators in disease-relevant models, including breast cancer models
- Combination approaches leveraging the spatiotemporal specificity of allosteric modulators with other targeted therapies
- Advanced delivery strategies to further enhance tissue specificity and reduce off-target effects
As our understanding of A1R pharmacology continues to evolve, these sophisticated targeting strategies offer renewed hope for harnessing the therapeutic potential of this important receptor in breast cancer and other disease contexts.
The Promise of Allosteric Modulators for Fine-Tuning A1R Activity
The adenosine A1 receptor (A1R), a class A G protein-coupled receptor (GPCR), has emerged as a promising therapeutic target in breast cancer research. Adenosine, its endogenous ligand, accumulates in tumor microenvironments and exerts diverse physiological effects through four receptor subtypes (A1, A2A, A2B, and A3) [60] [10]. The A1R is notably overexpressed in various breast cancer cell lines and participates in a regulatory feed-forward loop with estrogen receptor-α (ERα), a critical driver in hormone-responsive breast cancers [2] [41]. This relationship establishes A1R not merely as a passive target but as an active regulator of oncogenic signaling, making it a compelling subject for targeted therapeutic intervention.
Traditional drug discovery efforts focusing on orthosteric ligands (agonists/antagonists that bind the native adenosine site) have faced significant challenges. The high conservation of the orthosteric binding pocket across adenosine receptor subtypes hampers the achievement of sufficient subtype selectivity, leading to off-target effects [60] [61]. Furthermore, systemic activation or inhibition of A1R can cause dose-limiting on-target adverse effects, including bradycardia, atrioventricular block, and sedation [10]. These limitations have spurred investigation into allosteric modulation as a sophisticated strategy to fine-tune A1R activity with enhanced specificity and a superior therapeutic window, particularly in the complex context of breast cancer biology.
The Allosteric Advantage in A1R Targeting
Fundamental Concepts of Allosteric Modulation
Allosteric modulators are ligands that bind to a site on the receptor that is topographically distinct from the orthosteric site. They regulate receptor function by modulating the binding affinity and/or efficacy of the orthosteric ligand. Positive allosteric modulators (PAMs) enhance the response of the receptor to the endogenous agonist, while negative allosteric modulators (NAMs) attenuate it [60] [10]. Other classes include neutral allosteric ligands (NAL) and allosteric agonists, which can activate the receptor independently [60].
This mechanism offers several pharmacological advantages crucial for cancer therapy:
- Subtype Selectivity: Allosteric sites are under less evolutionary pressure to be conserved, enabling the development of highly subtype-selective compounds [60].
- Spatio-Temporal Specificity: PAMs augment the receptor's response only where and when the endogenous agonist (adenosine) is present. Since adenosine levels are significantly elevated in hypoxic tumor microenvironments, PAMs can exert context-dependent activity, potentially minimizing effects on healthy tissues [60] [10].
- Probe Dependence and Biased Signaling: Allosteric modulators can be engineered to promote specific, therapeutically beneficial signaling pathways (biased modulation) while avoiding those that lead to adverse effects [60].
- Ceiling Effect: The modulation effect of PAMs often reaches a plateau, providing a built-in safety mechanism that prevents over-activation of the receptor system [10].
Structural Basis of A1R Allosteric Modulation
Recent structural biology breakthroughs have illuminated the molecular details of A1R allosteric modulation. A seminal cryo-EM structure (PDB ID: 7LD3) revealed the binding of a PAM, MIPS521, within an extrahelical lipid-facing allosteric pocket formed by transmembrane helices 1, 6, and 7 [62]. This pocket is distinct from the orthosteric adenosine-binding site and the Gi protein interface.
Computational studies using Gaussian accelerated molecular dynamics (GaMD) simulations have further detailed the binding process. These simulations showed that prototypical PAMs like PD81723 and VCP171 spontaneously bind to a putative allosteric site on the receptor's extracellular loop 2 (ECL2) [61]. The binding of a PAM to this site stabilizes the entire receptor complex, including the orthosteric agonist, reducing its conformational freedom and dissociation rate, thereby enhancing and prolonging its effect [61]. This structural understanding provides a robust foundation for rational drug design.
Figure 1: Allosteric Modulation of the A1 Receptor
A1R in Breast Cancer Pathogenesis
The A1R-ERα Feed-Forward Loop
In hormone-responsive breast cancer, a critical mechanistic link exists between A1R and estrogen receptor-α (ERα). Research has demonstrated that A1R serves a dual role as both a target and a regulator of ERα, creating a short feed-forward loop that promotes cancer growth [2] [41].
- ERα Regulates A1R Expression: In ERα-positive MCF-7 breast cancer cells, estradiol (E2) upregulates Adora1 (the gene encoding A1R) mRNA and protein levels in a time- and concentration-dependent manner. This effect is reversed by the ERα antagonist ICI 182,780, confirming the dependence on ERα signaling [41].
- A1R Regulates ERα Activity and Proliferation: Intriguingly, silencing Adora1 using siRNA or inhibiting it with the selective antagonist DPCPX leads to a significant decrease in both ERα mRNA and protein levels. This ablation consequently reduces estrogen-responsive element (ERE)-dependent transcriptional activity and inhibits the binding of ERα to promoters of its target genes (e.g., TFF1) [2] [41]. Functionally, Adora1 knockdown or pharmacological inhibition reduces basal and E2-dependent proliferation in MCF-7 cells, while its overexpression in an ERα-negative cell line induces proliferation [41].
This reciprocal relationship positions A1R as a key node in a pro-proliferative signaling network, validating its pursuit as a therapeutic target in hormone-dependent breast cancer.
Pro-Apoptotic Effects of A1R Antagonism
Beyond its interplay with ERα, A1R signaling directly influences apoptotic pathways in breast cancer cells. A study on MCF-7 cells demonstrated that the selective A1R antagonist DPCPX significantly induced apoptosis and reduced cell viability, particularly 72 hours after treatment [5]. This pro-apoptotic effect was accompanied by a dramatic up-regulation in the expression of the tumor suppressor p53 and the executioner caspases 3, 8, and 9. Conversely, the A1R agonist CPA increased cell viability and reduced apoptosis, concurrently down-regulating p53 and caspase expression [5]. These findings indicate that tonic A1R activation provides a pro-survival signal in breast cancer cells and that its blockade can trigger programmed cell death.
Table 1: Experimental Evidence Linking A1R to Breast Cancer Pathogenesis
| Experimental Manipulation | Effect on ERα | Effect on Apoptosis Markers | Effect on Cell Proliferation | Key Citation |
|---|---|---|---|---|
| A1R Antagonism (DPCPX) | Decreased ERα protein levels [41] | Upregulated p53, Caspases 3, 8, 9 [5] | Reduced proliferation [41] | [5] [41] |
| A1R Agonism (CPA) | Information Not Provided | Downregulated p53 and caspases; Reduced apoptosis [5] | Increased cell viability [5] | [5] |
| siRNA Knockdown of A1R | Decreased ERα mRNA and protein [41] | Information Not Provided | Reduced basal and E2-dependent proliferation [41] | [41] |
| A1R Overexpression | Information Not Provided | Information Not Provided | Induced proliferation in ERα-negative cells [41] | [41] |
Experimental Protocols for A1R Research in Breast Cancer
Computational Screening for A1R-Targeted Compounds
Modern drug discovery leverages integrated bioinformatics and computational chemistry approaches to identify critical targets and design potent compounds.
Methodology Overview:
- Initial Compound Screening: A diverse set of compounds with known inhibitory effects on breast cancer cell lines (e.g., MCF-7, MDA-MB) is selected from literature. 3D quantitative structure-activity relationship (3D-QSAR) analyses are performed to evaluate spatial diversity and generate distinct conformers [9].
- Pharmacophore Modeling & Target Prediction: Pharmacophore models are constructed based on binding information to identify key structural features influencing biological activity. The chemical structures of potent compounds are used as input in the SwissTargetPrediction database to identify potential protein targets in Homo sapiens [9].
- Target Intersection Analysis: An online tool (e.g., Venny) is used to conduct an intersection analysis of the predicted targets for multiple active compounds. This helps identify shared targets, such as the A1R, which can be further validated through databases like PubChem using keywords "MDA-MB and MCF-7" [9].
- Molecular Docking: A ligand library is created using software like Discovery Studio. Docking simulations (e.g., with CHARMM) are performed to refine ligand shapes and charge distribution, analyzing binding interactions between candidate compounds and the A1R. Poses are selected based on high LibDock scores (e.g., >130) [9].
- Molecular Dynamics (MD) Simulations: The stability of the docked complexes is studied using software like GROMACS. Protein structures are optimized with a force field (e.g., AMBER99SB-ILDN), and ligands are parameterized with tools like ACPYPE (GAFF force field). The system is hydrated and neutralized, followed by energy minimization and restrained equilibration. Finally, unrestricted MD simulations (e.g., 15 ns) are run under isothermal-isobaric conditions to analyze binding dynamics [9].
In Vitro Validation of Anti-Tumor Efficacy
Following computational design, compounds must be validated in biological assays.
Cell-Based Proliferation and Apoptosis Assays:
- Cell Line: The ERα-positive MCF-7 human breast cancer cell line is a standard model [9] [5] [41].
- Compound Treatment: Cells are treated with novel candidate compounds (e.g., a rationally designed molecule like "Molecule 10" from recent research), positive controls (e.g., 5-Fluorouracil, 5-FU), and vehicle controls [9].
- Viability and IC50 Determination: Cell viability is measured using assays like MTT or MTS after treatment. The half-maximal inhibitory concentration (IC50) is calculated, with potent compounds showing values significantly lower than controls (e.g., 0.032 µM for a novel compound vs. 0.45 µM for 5-FU) [9].
- Apoptosis Analysis:
- Gene Expression: mRNA is extracted from treated cells (e.g., with A1R agonist CPA or antagonist DPCPX). The expression of p53 and caspases (3, 8, 9) is quantified using real-time PCR [5].
- Protein Analysis: Protein levels of ERα, proliferating cell nuclear antigen (PCNA), and A1R itself can be quantified via immunoblotting (Western blot) to confirm knockdown or functional effects [41].
- Apoptosis Rate: Direct measurement of apoptosis can be performed using flow cytometry with Annexin V/propidium iodide staining [5].
Figure 2: Integrated Workflow for A1R-Targeted Drug Discovery
The Scientist's Toolkit: Key Research Reagents
Table 2: Essential Reagents for A1R Research in Breast Cancer
| Reagent / Tool | Category | Function in Research | Example Use Case |
|---|---|---|---|
| DPCPX | Selective A1R Antagonist | Inhibits A1R signaling to study loss-of-function effects; validates target engagement. | Induces apoptosis in MCF-7 cells; reduces ERα levels and cell proliferation [5] [41]. |
| CPA (N⁶-Cyclopentyladenosine) | Selective A1R Agonist | Activates A1R signaling to study gain-of-function and pro-survival effects. | Increases MCF-7 cell viability and suppresses p53/caspase expression [5]. |
| siRNA/shRNA for Adora1 | Genetic Tool | Knocks down A1R gene expression to confirm phenotypic specificity. | Validates role of A1R in ERα regulation and proliferation independent of pharmacology [41]. |
| PAMs (e.g., PD81723, VCP171, T-62) | Positive Allosteric Modulator | Enhances endogenous adenosine signaling at A1R; tool for studying allosteric mechanisms. | Used in structural studies (MD simulations) and preclinical pain models; clinical trials for neuropathic pain [60] [61]. |
| MCF-7 Cell Line | Biological Model | ERα-positive human breast cancer cell line expressing A1R. | Standard in vitro model for studying A1R-ERα crosstalk and compound efficacy [9] [5] [41]. |
| A1R Antibodies | Detection Tool | Detects A1R protein expression and localization via Western blot, immunofluorescence. | Confirms A1R protein up-regulation by E2 and successful knockdown in experiments [41]. |
The investigation of allosteric modulators for the adenosine A1 receptor represents a paradigm shift in the approach to targeting this receptor for breast cancer therapy. The compelling biological rationale—centered on the A1R-ERα feed-forward loop and the pro-survival role of A1R—strongly supports its relevance in oncology [2] [41]. The allosteric advantage offers a path to overcome the historical hurdles of orthosteric ligands, promising unprecedented subtype selectivity and context-dependent activity aligned with the pathological tumor microenvironment [60] [10] [61].
Future work in this field will likely focus on translating structural insights into optimized drug candidates. The precise allosteric binding pockets revealed by cryo-EM and GaMD simulations provide a blueprint for structure-based drug design [62] [61]. Furthermore, exploring the potential of allosteric modulators to induce biased signaling—promoting anti-tumor pathways while avoiding those leading to adverse effects—remains a fertile area for research. As the first A1R PAMs, such as T-62, have already progressed to Phase II clinical trials for other indications (e.g., neuropathic pain), the foundational safety and efficacy data can inform their repurposing and optimization for oncology [60] [10]. Integrating A1R allosteric modulators with existing standard-of-care treatments, such as endocrine therapy for ER+ breast cancer, could pave the way for novel, effective, and finely-tuned combination therapies.
Optimizing Binding Affinity and Efficacy through Structure-Activity Relationship (SAR) Studies
Structure-Activity Relationship (SAR) studies serve as a fundamental pillar in modern drug discovery, enabling the rational optimization of lead compounds by systematically exploring how chemical structure modifications affect biological activity. Within the context of the preliminary investigation of adenosine A1 receptor (A1R) in breast cancer research, SAR provides the methodological framework to design ligands with enhanced binding affinity, subtype selectivity, and desired efficacy profiles [63]. The A1R, a class A G protein-coupled receptor (GPCR), represents a promising therapeutic target, though its drug development has been hampered by the challenge of separating therapeutic effects from adverse side effects such as bradycardia and sedation [63] [64]. SAR studies are crucial for overcoming these hurdles by informing the design of ligands that can engage the receptor in novel ways, potentially through allosteric modulation or biased agonism, to achieve a refined pharmacological response [65] [64].
Recent evidence suggests the A1R plays a significant role in tumor biology. In breast cancer cell lines, A1R has been identified as a shared target of several anticancer compounds, and its modulation can influence apoptosis and proliferation [9] [66]. For instance, a novel molecule (Molecule 10) developed through SAR-guided optimization demonstrated potent antitumor activity against MCF-7 breast cancer cells with an IC₅₀ of 0.032 µM, significantly outperforming the positive control 5-FU [9]. This underscores the potential of targeting A1R in oncology and highlights the critical importance of meticulous SAR analysis in developing effective and safe therapeutic agents.
Key Structural Modifications and Their SAR in A1R Ligands
The endogenous ligand for A1R is adenosine, which consists of an adenine moiety and a ribose sugar. SAR studies have extensively explored modifications at various positions on this scaffold to enhance affinity, selectivity, and metabolic stability. The key sites for chemical modification are the N⁶-position on the adenine ring, the C2-position, and the ribose moiety, particularly the 5'-position [67] [68] [69].
Table 1: Key Structural Modifications and Their Effects on A1R Activity
| Modification Site | Structural Change | Effect on Binding Affinity | Effect on Selectivity & Efficacy |
|---|---|---|---|
| N⁶-Position | Cycloalkyl (e.g., cyclopentyl) | Increases affinity [67] | Enhances A1R selectivity over other subtypes [67] |
| Benzyloxycyclopentyl (e.g., BnOCPA) | High potency retained [68] | Confers high A1R selectivity and unique G protein signaling bias [68] | |
| Phenoxycyclopentyl | High affinity with halogen substituents [67] | ∼1500-fold A1R selectivity for optimized NECA-based compounds [67] | |
| C2-Position | Chloro (Cl) | Enhances affinity at A1R and A3R [69] | Can reduce intrinsic efficacy, leading to partial agonism [69] |
| Cyano (CN) | Can enhance affinity, depending on N⁶-substituent [69] | Reduces efficacy; small groups preferred for high efficacy [69] | |
| 5'-Position (Ribose) | N-ethylcarboxamido (NECA) | Generally increases potency [67] | Can reduce A1R selectivity compared to adenosine-based analogs [68] |
| Bitopic Ligand (VCP746) | Dramatically higher affinity (KI ∼60 nM) than adenosine [64] | Introduces biased signaling, separating cardioprotection from bradycardia [64] |
N⁶-Position Substitutions
The N⁶-position is one of the most widely modified sites to achieve high A1R affinity and selectivity. Introducing hydrophobic substituents like the cyclopentyl group (as in CPA) is a classic strategy to boost A1R affinity [67]. Extending this further by appending a benzyloxy or phenoxy group to the N⁶-cyclopentyl ring (e.g., BnOCPA) can result in compounds with exceptional A1R selectivity and a unique pharmacological profile [68]. The structure of the aromatic ring is critical; a halogen atom in the meta position (e.g., bromine) often confers high affinity for the A1R [67]. Molecular modeling suggests this group engages a hydrophobic subpocket within the receptor, contributing to the observed selectivity [67].
C2-Position and Ribose Modifications
Substitution at the C2-position of the adenine ring can significantly modulate both affinity and intrinsic efficacy. While a 2-chloro substitution is known to enhance the affinity of many adenosine derivatives, other small groups like cyano (CN) can also be beneficial [69]. The effect is context-dependent, varying with the specific N⁶-substituent present. A key SAR finding is that certain 2-substitutions can reduce intrinsic efficacy, potentially converting a full agonist into a partial agonist, which offers a pathway to fine-tune functional output [69]. Modification of the ribose 5'-position to a N-ethylcarboxamido group (yielding NECA) typically increases potency. However, NECA-based compounds can display different selectivity profiles compared to their adenosine-based counterparts, a critical consideration during optimization [67] [68].
Advanced Ligand Design: Allosteric and Bitopic Modulation
Moving beyond traditional orthosteric targeting, advanced SAR strategies are exploring allosteric and bitopic modulation to achieve unprecedented selectivity and control over receptor signaling.
Allosteric Modulators
Positive Allosteric Modulators (PAMs), such as MIPS521, bind to a site topographically distinct from the orthosteric adenosine binding pocket [63] [65]. They enhance the affinity and/or efficacy of the endogenous ligand, offering several advantages: they are active only when and where adenosine is released, providing spatial and temporal selectivity, and they can exhibit greater subtype selectivity due to lower conservation of allosteric sites [63]. This mechanism is being explored for non-opioid analgesics and other conditions to avoid the side effects of direct orthosteric agonists [65].
Bitopic Ligands
Bitopic ligands represent a sophisticated SAR-driven approach that hybridizes orthosteric and allosteric pharmacophores into a single molecule [64]. A prime example is VCP746, which links adenosine to a PAM (VCP171) via a flexible linker. The SAR of the linker is critical; a bell-shaped relationship with linker length is observed, with a 6-carbon linker (in VCP746) yielding the highest affinity [64]. This bitopic design results in a ligand that binds concomitantly to both the orthosteric and allosteric sites, a mode that can stabilize unique receptor conformations and drive biased signaling. This has been successfully demonstrated with VCP746, which provides cardioprotection in vitro without inducing bradycardia in isolated rat atria, effectively separating on-target efficacy from on-target adverse effects [64].
Experimental Protocols for Key A1R SAR Studies
In Vitro Binding and Functional Assays
Radioligand Binding Assay is used to determine the affinity (Kᵢ) of novel compounds for the A1R.
- Protocol: Membranes from cells expressing recombinant human A1R (e.g., CHO-K1, HEK293) or native tissue (e.g., rat brain cortex) are prepared. These membranes are incubated with a known concentration of a radiolabeled antagonist (e.g., [³H]DPCPX) and increasing concentrations of the test compound. Non-specific binding is defined by a high concentration of a standard agonist (e.g., R-PIA). Bound and free radioligand are separated by vacuum filtration. Data are analyzed using nonlinear regression to determine the IC₅₀ and subsequently the Kᵢ of the test compound [67] [70].
Functional Assay – cAMP Accumulation: Since A1R is primarily Gᵢ/o-coupled, its activation inhibits adenylate cyclase and reduces intracellular cAMP levels.
- Protocol: Cells expressing A1R are stimulated with forskolin (to elevate cAMP) and co-treated with a range of concentrations of the test agonist. After cell lysis, cAMP content is quantified using a competitive binding or immunoassay method. The concentration-response data are fit to a curve to determine the agonist's EC₅₀ and intrinsic activity (efficacy relative to a reference full agonist) [67] [68].
NanoBRET Binding Assay: A live-cell assay used to measure both affinity and binding kinetics (kon and koff).
- Protocol: Cells expressing a NanoLuc-fused A1R are incubated with a tracer ligand and test compounds. The energy transfer signal is monitored over time. By analyzing association and dissociation curves, the kinetic rate constants and affinity of the test compound can be quantified [67].
Molecular Modeling and Dynamics
Molecular Docking predicts the binding pose of a ligand within the A1R binding site.
- Protocol: A high-resolution structure of A1R (e.g., PDB ID: 7LD3) is prepared by adding hydrogen atoms and optimizing side-chain conformations. The 3D structure of the ligand is energy-minimized. Docking simulations (e.g., using CHARMM) are performed to generate plausible binding poses, which are scored based on complementary interactions (e.g., LibDock score). Poses with scores >130 are typically considered high-affinity candidates [9].
Molecular Dynamics (MD) Simulations assess the stability of the docked protein-ligand complex under near-physiological conditions.
- Protocol: The docked complex is solvated in a water box (e.g., TIP3P model) and neutralized with ions. The system is energy-minimized and equilibrated. Production simulations are run for tens to hundreds of nanoseconds (e.g., using GROMACS with the AMBER99SB-ILDN force field). Trajectories are analyzed for root-mean-square deviation (RMSD), binding interactions, and residue fluctuations to evaluate complex stability and mechanism of action [9] [68].
A1R Signaling Pathways and Ligand Effects
The A1R signals primarily through Gᵢ/o proteins, leading to the inhibition of adenylate cyclase and a decrease in intracellular cAMP levels [63] [24]. However, it also promiscuously couples to other effector systems, including the activation of potassium channels, inhibition of calcium channels, and stimulation of phospholipase C and ERK1/2 MAP kinase pathways [24]. This multiplicity of signaling pathways underlies the concept of biased agonism, where different ligands can stabilize distinct receptor conformations, preferentially activating a subset of these pathways [65] [64].
For example, the bitopic ligand VCP746 and the highly selective agonist BnOCPA have been shown to exhibit biased signaling. BnOCPA specifically activates GαoB protein subtypes, translating into potent in vivo analgesia without sedation or bradycardia [68]. The PAM MIPS521 can also differentially modulate the G protein selectivity of adenosine and BnOCPA, suggesting that allosteric ligands can fine-tune the signaling of orthosteric ligands [65]. In breast cancer research, understanding and harnessing this bias could be key to developing A1R-targeted therapies that inhibit proliferation or induce apoptosis without triggering detrimental side effects.
Table 2: Key Research Reagent Solutions for A1R SAR Studies
| Reagent / Resource | Function / Application | Example(s) |
|---|---|---|
| Cell Lines | In vitro models for binding and functional assays. | CHO-K1, HEK293 stably expressing human A1R; MCF-7, MDA-MB breast cancer cells [9] [67]. |
| Radioligands | Quantitative measurement of ligand affinity in binding assays. | [³H]DPCPX (antagonist), [¹²⁵I]I-AB-MECA (agonist), [³H]R-PIA (agonist) [67] [70]. |
| Reference Ligands | Standards for determining selectivity, efficacy, and potency. | Agonists: CPA, NECA, CCPA, BnOCPA. Antagonists: DPCPX, XAC [67] [68] [70]. |
| cAMP Assay Kits | Measurement of functional activity via cAMP modulation. | Commercially available kits based on competitive binding, ELISA, or BRET/FRET technologies [67]. |
| Molecular Modeling Software | Visualization, docking, and dynamics simulations. | VMD (visualization), GROMACS (MD simulations), CHARMM (force field/docking) [9]. |
| A1R Crystal/Cryo-EM Structures | Templates for structure-based drug design. | PDB ID: 7LD3 (A1R bound to adenosine, Gi2, and allosteric modulator) [9] [68]. |
SAR studies remain an indispensable component of the drug discovery pipeline for the adenosine A1 receptor. The systematic exploration of chemical space around the adenosine scaffold has yielded ligands with exceptional affinity and selectivity, while also paving the way for novel pharmacological approaches like allosteric and bitopic modulation. The preliminary investigation of A1R in breast cancer provides a compelling therapeutic context for these efforts [9] [66]. The future of A1R SAR lies in the deeper integration of computational and experimental methods, the continued pursuit of biased signaling to separate therapeutic effects from side effects, and the translation of these findings into validated in vivo disease models. As structural biology techniques provide ever more detailed views of receptor-ligand interactions, the potential for rational, SAR-driven design of next-generation A1R therapeutics for cancer and other diseases is greater than ever.
The adenosinergic pathway represents a critical signaling network within the tumor microenvironment, characterized by remarkable biochemical redundancy and complex interdependencies among its constituent enzymes. This pathway exerts profound immunosuppressive and tumor-promoting effects, with particular significance in hormone-dependent cancers such as breast cancer. This technical review examines the intricate web of adenosine-generating and adenosine-degrading enzymes, focusing on their collective impact on cancer progression and their emerging role as a promising therapeutic target. Within the specific context of breast cancer, we explore the preliminary investigation of the adenosine A1 receptor (A1R) as a key mediator of estrogen receptor-alpha (ERα) signaling and cellular proliferation, providing a mechanistic framework for future therapeutic interventions.
Adenosine is an endogenous purine nucleoside that functions as a key signaling molecule in both physiological and pathophysiological conditions [71]. Under normal cellular conditions, extracellular adenosine concentrations are maintained at approximately 300 nM, but these levels can rapidly increase to 600-1200 nM during cellular stress, such as hypoxia, ischemia, or inflammation within the tumor microenvironment [72]. The adenosinergic pathway encompasses the complete network of enzymes, transporters, and receptors that collectively regulate the production, degradation, and signaling of adenosine.
In oncology, this pathway has gained significant attention as a major mediator of tumor immune evasion and cancer progression [73] [74]. The accumulation of extracellular adenosine within the tumor microenvironment creates a potent immunosuppressive barrier that limits antitumor immunity and promotes angiogenesis, metastasis, and therapeutic resistance [73]. The pathway's complexity is further heightened in hormone-responsive cancers like breast cancer, where emerging evidence demonstrates crosstalk between adenosinergic signaling and hormone receptor pathways, particularly involving the adenosine A1 receptor [6].
Core Components of the Adenosinergic Pathway
Adenosine-Generating Enzymes
The production of extracellular adenosine occurs through multiple complementary and redundant enzymatic pathways, ensuring robust generation of this immunosuppressive metabolite despite potential inhibition of individual components.
Table 1: Major Adenosine-Generating Enzyme Systems
| Enzyme System | Key Components | Catalytic Function | Tumor Expression |
|---|---|---|---|
| CD39-CD73 Axis | CD39 (ENTPD1), CD73 (NT5E) | ATP → ADP → AMP → Adenosine | Upregulated in multiple cancers [73] |
| Alternative Pathway | CD38, CD203a (ENPP3), CD73 | NAD+ → ADPR → AMP → Adenosine | Active in hypoxic conditions [73] [71] |
| Alkaline Phosphatase Pathway | Tissue-nonspecific ALP (TNAP) | ATP → Adenosine + 3Pi | Broad substrate specificity [73] |
| Nucleotide Recycling | Adenylate kinase (AK1), Nucleoside diphosphate kinase (NME1/2) | ATP + AMP 2ADP; ATP + NDP ADP + NTP | Maintains purine pool homeostasis [74] |
The CD39-CD73 axis represents the most characterized pathway for extracellular adenosine production [73]. CD39 (ecto-nucleoside triphosphate diphosphohydrolase-1) initiates the process by hydrolyzing extracellular ATP (eATP) or ADP to AMP, which is subsequently converted to adenosine by CD73 (ecto-5'-nucleotidase) [73] [74]. This pathway is particularly significant in the tumor microenvironment where hypoxic conditions stimulate ATP release from stressed and dying cells.
Alternative pathways provide redundancy in adenosine generation. The CD38-CD203a-CD73 axis becomes particularly important in hypoxic conditions, where CD38 utilizes NAD+ as a substrate to generate adenosine diphosphate ribose (ADPR), which is subsequently converted to AMP by CD203a and finally to adenosine by CD73 [73] [71]. This pathway may compensate when CD39 activity is limited. Additionally, tissue-non-specific alkaline phosphatases (TNAPs) and prostatic acid phosphatases (PAPs) can directly hydrolyze nucleotides to adenosine, further expanding the enzymatic repertoire available for adenosine production [73].
Notably, ATP can be regenerated in the extracellular space through the actions of adenylate kinase (AK) and nucleoside diphosphate kinase (NDPK), which maintain local purine pools and provide sustained substrate availability for ectoenzymes like CD39 and CD203a [73] [74]. This complex network of interconnected enzymes creates a robust system that maintains adenosine signaling even when individual components are compromised.
Adenosine Receptors and Signaling
Extracellular adenosine mediates its diverse physiological effects through four G-protein-coupled receptors: A1, A2A, A2B, and A3 receptors [71] [72]. These receptors exhibit distinct affinities for adenosine and couple to different intracellular signaling pathways.
Table 2: Adenosine Receptor Characteristics
| Receptor | Adenosine Affinity | G-protein Coupling | Primary Signaling Effect | Expression in Breast Cancer |
|---|---|---|---|---|
| A1R | High (Ki ≈ 310 nM) [73] | Gi/o [72] | ↓ cAMP, ↑ K+ efflux [72] | Upregulated by E2, mediates proliferation [6] [54] |
| A2AR | High (Ki ≈ 700 nM) [73] | Gs/Golf [71] | ↑ cAMP, anti-inflammatory [72] | Upregulated by E2 [54] |
| A2BR | Low (Ki ≥ 10 μM) [73] | Gs, Gq [71] [72] | ↑ cAMP, ↑ phospholipase activity [71] | Downregulated by E2 at high doses [54] |
| A3R | Context-dependent [73] | Gi/o, Gq [71] [72] | ↓ cAMP, ↑ phospholipase activity [71] | Upregulated by E2 [54] |
The A1 receptor (A1R) couples to Gi/o proteins, leading to inhibition of adenylate cyclase and reduced intracellular cAMP levels [72]. In cardiovascular tissues, A1R activation causes cell hyperpolarization through increased K+ efflux and inhibits Ca2+ current, explaining its anti-arrhythmic effects [72]. In breast cancer, A1R has emerged as a particularly significant mediator, with studies demonstrating its regulation by estrogen receptor-alpha and its essential role in mediating the proliferative effects of estradiol [6].
The A2A receptor (A2AR) primarily couples to Gs proteins, stimulating adenylate cyclase and increasing intracellular cAMP [71]. This signaling generally produces anti-inflammatory effects and serves as a feedback mechanism to limit excessive immune activation [72]. The A2B receptor (A2BR) requires higher adenosine concentrations for activation (in the micromolar range) and can couple to both Gs and Gq proteins, enabling diverse signaling outcomes including increased cAMP and phospholipase activity [73] [71]. The A3 receptor (A3R) exhibits context-dependent affinity for adenosine and can form heterodimers with other adenosine receptors, particularly A2A, potentially altering their pharmacological profiles [73].
Adenosine Clearance and Metabolism
The termination of adenosine signaling involves multiple mechanisms, including cellular uptake and enzymatic degradation. Equilibrative nucleoside transporters (ENTs) and concentrative nucleoside transporters (CNTs) mediate adenosine uptake into cells [73] [74]. Once internalized, adenosine can be phosphorylated to AMP by adenosine kinase (ADK) or deaminated to inosine by adenosine deaminase (ADA) [74] [75]. extracellular adenosine can also be directly deaminated by ecto-ADA, resulting in the production of inosine, which is further converted to hypoxanthine by purine nucleoside phosphorylase (PNP) [73]. This multi-layered system for adenosine clearance ensures precise spatiotemporal control over adenosine signaling.
Redundancy and Interdependence in the Adenosinergic Pathway
The adenosinergic pathway exhibits remarkable redundancy, with multiple enzymatic routes capable of generating adenosine, ensuring reliable adenosine production even when individual components are compromised. This redundancy presents a significant challenge for therapeutic interventions targeting adenosine production.
The interdependence of pathway components is equally important. The CD38-CD203a-CD73 axis can compensate for reduced CD39 activity, particularly under hypoxic conditions where NAD+ availability increases [73] [71]. Similarly, alkaline phosphatases provide a bypass mechanism that can generate adenosine directly from ATP or other nucleotides when specialized nucleotidases are inhibited [73]. This functional overlap ensures that adenosine production remains robust across diverse tissue environments and pathological conditions.
The diagram below illustrates the complex network of enzymes, substrates, and products that constitute the adenosinergic pathway, highlighting the multiple redundant pathways for adenosine generation:
Figure 1: Redundant pathways for adenosine generation and metabolism. Multiple enzymatic routes can produce adenosine from various substrates, creating a robust network that maintains adenosine signaling despite inhibition of individual components.
Adenosine A1 Receptor in Breast Cancer: Preliminary Investigations
A1R as a Mediator of Estrogen Signaling
Emerging research has revealed a significant connection between the adenosine A1 receptor and estrogen receptor signaling in breast cancer. Studies using ERα-positive MCF-7 human breast cancer cells have demonstrated that estradiol (E2) upregulates A1R mRNA and protein levels in a dose-dependent manner, an effect that is reversible by the estrogen antagonist ICI 182,780 [6] [54]. This regulation appears to be specific to different adenosine receptor subtypes, with A1R, A2AR, and A3R showing upregulation in response to E2, while A2BR expression decreases at higher E2 concentrations [54].
Intriguingly, a feed-forward loop has been identified between E2, ERα, and A1R. While E2 upregulates A1R expression, ablation of A1R in turn decreases both mRNA and protein levels of ERα and consequently reduces estrogen-responsive element-dependent ERα transcriptional activity [6]. This reciprocal regulation suggests that A1R is not merely a passive target of estrogen signaling but an active participant required for full transcriptional activity of ERα upon estrogen stimulation.
A1R in Breast Cancer Proliferation
Functional studies have established the importance of A1R in breast cancer cell proliferation. Small interference RNA-mediated ablation of A1R in ERα-positive cells significantly reduces both basal and E2-dependent proliferation [6]. Conversely, A1R overexpression in ERα-negative cell lines induces proliferation, establishing its direct role in growth promotion [6]. Pharmacological inhibition using the selective A1R antagonist DPCPX similarly reduces proliferation, confirming A1R as a meaningful therapeutic target in hormone-dependent breast cancer [6].
The molecular mechanisms underlying A1R-mediated proliferation involve regulation of cell cycle progression. Knockdown of A1R affects G1 checkpoint control, leading to increased accumulation of cells in G2/M phase, which confirms its stimulatory effects on cell cycle progression [54]. Additionally, A1R ablation decreases the binding activity of ERα to the promoter of its target gene TFF1 (trefoil factor 1), leading to reduced TFF1 promoter activity and mRNA levels [6]. This suggests that A1R is required for the full transcriptional activity of ERα on estrogen stimulation.
Experimental Approaches for Adenosinergic Pathway Investigation
Research Reagent Solutions
Table 3: Essential Research Reagents for Adenosinergic Pathway Studies
| Reagent Category | Specific Examples | Research Application | Key Findings Enabled |
|---|---|---|---|
| A1R Agonists | N6-Cyclopentyladenosine (CPA) | A1R-specific activation | Validation of A1R-mediated proliferative signaling [6] |
| A1R Antagonists | DPCPX, 8-Phenyltheophylline | A1R pharmacological inhibition | Confirmation of A1R role in breast cancer proliferation [6] |
| ER Modulators | 17β-estradiol (E2), ICI 182,780 | Estrogen pathway manipulation | Demonstration of ERα-A1R feed-forward loop [6] [54] |
| Gene Silencing Tools | siRNA against ADORA1 | A1R knockdown studies | Elucidation of A1R effects on ERα transcriptional activity [6] |
| Cell Viability Assays | MTT assay | Proliferation measurement | Quantification of E2 and adenosine effects on cell growth [54] |
Methodological Framework
Experimental Protocol 1: Investigating A1R-ERα Interactions in Breast Cancer Cells
Cell Culture Conditions: Maintain MCF-7 cells (ERα-positive) in RPMI-1640 medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 mg/mL streptomycin at 37°C in 5% CO2 [54].
Estrogen Treatment: For E2 stimulation, culture cells in estrogen-depleted medium (charcoal-stripped serum) for 24 hours prior to treatment with 0.001-0.1 μM 17β-estradiol for 12 hours [54].
Receptor Antagonism Studies: Apply estrogen antagonist ICI 182,780 (1 μM) 1 hour prior to E2 treatment to confirm ERα-dependent effects [54].
Gene Expression Analysis: Extract total RNA using appropriate kits. Perform reverse transcription followed by quantitative real-time PCR using primers specific for adenosine receptor subtypes (A1, A2A, A2B, A3) with GAPDH as housekeeping gene. Calculate relative expression using the 2−ΔΔCt method [54].
Proliferation Assays: Seed 5 × 10^3 cells/well in 96-well plates. After treatment, add MTT compound (5 mg/mL final concentration) and incubate for 4 hours. Dissolve formazan crystals with DMSO and measure absorbance at 570 nm [54].
Experimental Protocol 2: Computational Screening for A1R-Targeted Compounds
Molecular Docking: Perform docking simulations between candidate compounds and the human adenosine A1 receptor-Gi2 protein complex (PDB ID: 7LD3) using Discovery Studio with CHARMM force field [9] [19].
Binding Assessment: Evaluate binding stability using LibDock scores, with scores >130 indicating strong binding affinity. Analyze absolute and relative energy values of complexes [19].
Molecular Dynamics Simulations: Conduct MD simulations using GROMACS with AMBER99SB-ILDN force field for proteins and GAFF for ligands. Use TIP3P water model in cubic boxes with minimum atom-box boundary distance of 0.8 nm [9] [19].
Simulation Parameters: Perform energy minimization, followed by 150 ps restrained MD at 298.15 K. Run unrestricted MD simulations for 15 ns with 0.002 ps time step under isothermal-isobaric conditions (298.15 K, 1 bar) [9].
Trajectory Analysis: Use VMD software to analyze motion trajectories, capturing data every 200 frames to document molecular binding processes and dynamic behavior [9].
The following diagram illustrates the integrated experimental workflow for investigating the A1R-ERα axis in breast cancer:
Figure 2: Integrated experimental workflow for investigating A1R-ERα axis in breast cancer. The approach combines experimental biology methods with computational screening to comprehensively evaluate A1R function and identify potential therapeutic compounds.
Therapeutic Implications and Future Directions
The redundant nature of the adenosinergic pathway presents significant challenges for therapeutic intervention, as targeting individual components may be insufficient to effectively disrupt adenosine-mediated immunosuppression and tumor promotion. Combination approaches that simultaneously target multiple pathway components or integrate adenosine pathway inhibition with existing therapies show greater promise.
In breast cancer, the interconnection between A1R and ERα signaling suggests potential therapeutic strategies that concurrently target both pathways. The development of selective A1R antagonists represents a promising approach, particularly in hormone-responsive breast cancers where A1R appears to augment ERα signaling [6]. Preclinical studies have demonstrated that rationally designed compounds targeting A1R can exhibit potent antitumor activity against MCF-7 cells with IC50 values as low as 0.032 μM, significantly outperforming conventional chemotherapeutic agents like 5-fluorouracil (IC50 = 0.45 μM) [9] [19].
Future research directions should focus on several key areas: (1) developing more comprehensive biomarkers for patient stratification based on adenosinergic pathway activation; (2) optimizing combination therapies that target multiple adenosine-generating enzymes simultaneously; (3) exploring the timing and sequencing of adenosine pathway inhibition relative to other treatment modalities; and (4) investigating the potential of targeting intracellular adenosine metabolism in addition to extracellular signaling. As our understanding of the complex adenosinergic network deepens, particularly its intersection with hormonal signaling in breast cancer, more effective therapeutic strategies will emerge to overcome the challenges posed by this robust and redundant pathway.
Validating A1R as a Therapeutic Target: Comparative Analysis and Preclinical Evidence
Comparative Analysis of A1R Antagonist Efficacy Against Standard Chemotherapeutics (e.g., 5-FU)
Within the broader context of a preliminary investigation of the adenosine A1 receptor (A1R) in breast cancer research, this whitepaper provides a comparative analysis of the therapeutic potential of A1R antagonists against standard chemotherapeutic agents, with a specific focus on 5-Fluorouracil (5-FU). The adenosine signaling pathway has emerged as a critical modulator of the tumor microenvironment, influencing cancer cell proliferation, survival, and response to therapy. In particular, the A1R subtype has been identified as being overexpressed in breast cancer cells, where it appears to play a role in promoting cell viability and mitigating apoptosis [4]. This analysis details the mechanistic basis, experimental efficacy, and protocol considerations for exploring A1R antagonism as a potential therapeutic strategy, providing a technical resource for researchers and drug development professionals engaged in oncology and cancer pharmacology.
Background and Mechanistic Rationale
The Role of Adenosine A1 Receptor in Breast Cancer
Adenosine receptors, including the A1 subtype, are G-protein coupled receptors (GPCRs) that are frequently found to be upregulated in various tumor cells [76]. In the context of breast cancer, evidence indicates that the A1 receptor functions as both a target and a regulator of estrogen receptor-alpha (ERα) action. Specifically, estradiol (E2) upregulates A1R mRNA and protein levels in ERα-positive breast cancer cells, an effect that can be reversed by the estrogen antagonist ICI 182,780 [6]. This establishes a feed-forward loop wherein E2/ERα signaling enhances A1R expression, and the A1R, in turn, is required for the full transcriptional activity of ERα upon E2 stimulation. Ablation of A1R reduces ERα levels and its transcriptional activity on target genes, underscoring a significant cross-talk between these pathways that favors breast cancer growth [6].
Mechanisms of Standard Chemotherapeutics: 5-Fluorouracil
5-Fluorouracil (5-FU) remains a cornerstone chemotherapeutic agent for a range of solid tumors, including breast cancer. Its cytotoxic mechanisms are twofold:
- Inhibition of Thymidylate Synthase (TS): This enzyme is critical for the de novo synthesis of thymidine, a nucleotide necessary for DNA replication. Inhibition of TS leads to thymidine depletion and impaired DNA synthesis.
- Misincorporation into RNA and DNA: Metabolites of 5-FU can be erroneously incorporated into RNA and DNA, disrupting normal nucleic acid function and leading to cell death [77].
Despite its widespread use, the efficacy of 5-FU is often limited by the development of chemoresistance. Mechanisms underlying 5-FU resistance are diverse and can include alterations in drug metabolism (e.g., changes in TS or dihydropyrimidine dehydrogenase expression), enhanced DNA repair, activation of survival pathways such as WNT/β-catenin and NOTCH signaling, and aberrations in cell cycle control and apoptosis [77]. Furthermore, the efficacy of 5-FU is known to rely on functional p53 signaling, and mutations in the p53 gene are a common contributor to resistance in breast cancer and other malignancies [78].
Comparative Efficacy Data: A1R Antagonists vs. 5-FU
Direct comparative studies on A1R antagonists and 5-FU are limited in the available literature. However, data from individual investigations can be synthesized to provide a preliminary comparison of their anti-proliferative and pro-apoptotic effects, particularly in the MCF-7 breast cancer cell line model.
Table 1: Comparative Efficacy of DPCPX (A1R Antagonist) and 5-FU in MCF-7 Breast Cancer Cells
| Parameter | DPCPX (A1R Antagonist) | 5-Fluorouracil (5-FU) |
|---|---|---|
| Primary Molecular Target | Adenosine A1 Receptor (A1R) | Thymidylate Synthase (TS); RNA/DNA |
| Reported IC₅₀ | 87 nM after 24 hours [4] | ~20 μM after 48 hours [77] |
| Effect on Apoptosis | Significant induction (Annexin V/PI assay) [4] | Induced at higher concentrations or longer durations [77] |
| Key Gene Expression Changes | Upregulation of p53, Caspase-3, -8, and -9 [4] | Efficacy relies on functional p53; mutations confer resistance [78] |
| Effect on Cell Viability | Reduced in a time-dependent manner (MTT assay) [4] | Reduced, but resistance develops via multiple pathways [77] |
| Proposed Mechanism in MCF-7 | Antagonism of pro-survival A1R signaling, leading to p53-mediated caspase activation [4] | Inhibition of DNA/RNA synthesis; cell cycle disruption; p53-dependent apoptosis [77] [78] |
The data suggests that the A1R antagonist DPCPX operates through a distinct, receptor-mediated mechanism that culminates in the activation of classic tumor suppressor and apoptotic pathways. Its potency in the nanomolar range, as indicated by the IC₅₀ value, highlights a potentially potent therapeutic effect. The ability of DPCPX to induce apoptosis even in cells with wild-type p53 is a key differentiator, though its efficacy in p53-mutant models requires further investigation.
Experimental Protocols for Key Assays
To facilitate the replication and further investigation of these findings, detailed methodologies for core experiments are outlined below.
Cell Culture and Treatment
- Cell Line: MCF-7 human breast cancer cells (ERα+, wild-type p53).
- Culture Conditions: Maintain cells in Dulbecco's Modified Eagle Medium (DMEM/F12) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin. Incubate at 37°C in a humidified atmosphere with 5% CO₂.
- Test Compounds: Prepare stock solutions of the A1R antagonist DPCPX and agonist CPA in dimethyl sulfoxide (DMSO). Prepare 5-FU in a suitable solvent (e.g., saline or DMSO). Dilute all working concentrations in culture medium immediately before use, ensuring the final concentration of DMSO does not exceed 0.1% (v/v) to avoid cytotoxicity.
- Treatment Regimen: Seed cells at an appropriate density (e.g., 1 x 10⁴ cells/well for 24-well plates) and allow to adhere for 24 hours. Replace medium with fresh medium containing the desired concentrations of DPCPX, CPA, or 5-FU. Include vehicle control (e.g., 0.1% DMSO). Perform treatments in triplicate and conduct experiments at least three times independently [4].
MTT Cell Viability Assay
This colorimetric assay measures the metabolic activity of cells as a surrogate for viability.
- Cell Seeding and Treatment: Seed MCF-7 cells in 24-well plates and treat as described in Section 4.1 for desired time intervals (e.g., 24, 48, 72h).
- MTT Incubation: At each time point, add MTT reagent (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) to each well to a final concentration of 0.5 mg/mL. Incubate the plate for 3-4 hours at 37°C.
- Solubilization: Carefully remove the medium and add DMSO to each well to solubilize the formed purple formazan crystals.
- Absorbance Measurement: Transfer 100 µL of the solubilized solution to a 96-well plate and measure the optical density (OD) at a wavelength of 570 nm using a microplate reader. Calculate the percentage of cell viability relative to the vehicle control [4].
Flow Cytometry for Apoptosis Detection
Quantify apoptotic cells using Annexin V-FITC and Propidium Iodide (PI) staining.
- Cell Harvesting: After treatment, harvest both adherent and floating cells by trypsinization and combine them by centrifugation.
- Staining: Wash the cell pellet with cold phosphate-buffered saline (PBS). Resuspend approximately 4 x 10⁵ cells in 195 μL of 1X binding buffer. Add 5 μL of Annexin V-FITC and 10 μL of PI solution (20 μg/mL). Gently vortex the cells and incubate for 10-15 minutes at room temperature in the dark.
- Analysis: Within 1 hour, analyze the stained cells using a flow cytometer. Distinguish cell populations as follows: viable cells (Annexin V⁻/PI⁻), early apoptotic (Annexin V⁺/PI⁻), late apoptotic (Annexin V⁺/PI⁺), and necrotic (Annexin V⁻/PI⁺) [4].
Gene Expression Analysis by Quantitative Real-Time PCR (qRT-PCR)
Evaluate the mRNA expression of genes involved in apoptosis.
- RNA Extraction: At designated time points post-treatment, extract total RNA from cells using a commercial kit (e.g., RNeasy Mini Kit), including an on-column DNase I digestion step to remove genomic DNA contamination.
- cDNA Synthesis: Reverse-transcribe 100 ng of total RNA into complementary DNA (cDNA) using a reverse transcription kit (e.g., RevertAid First Strand cDNA Synthesis Kit) with oligo(dT) or random hexamer primers.
- qPCR Amplification: Perform real-time PCR reactions using a SYBR Green master mix. Use specific primers for p53, caspases-3, -8, -9, and a housekeeping gene (e.g., GAPDH or β-actin) for normalization. Run reactions in triplicate on a real-time PCR system.
- Data Analysis: Calculate the relative gene expression using the comparative CT (2^(-ΔΔCT)) method [4].
Signaling Pathways and Workflows
The following diagrams, generated using DOT language, illustrate the proposed signaling pathways and experimental workflows central to this comparative analysis.
A1R Antagonist-Mediated Apoptotic Signaling Pathway
A1R Antagonist Mechanism
5-FU Mechanism and Resistance Pathways
5-FU Mechanism and Resistance
Experimental Workflow for Comparative Analysis
Experimental Workflow
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Research Reagents for Investigating A1R in Breast Cancer
| Reagent / Tool | Function / Application | Example / Note |
|---|---|---|
| Selective A1R Antagonist | Pharmacological inhibition of A1R to assess its role in cell survival and proliferation. | DPCPX (1,3-dipropyl-8-cyclopentylxanthine); used at nanomolar concentrations (e.g., 87 nM) [4]. |
| Selective A1R Agonist | Pharmacological activation of A1R; serves as a control to confirm antagonist effects. | CPA (N⁶-Cyclopentyladenosine) [4]. |
| 5-Fluorouracil (5-FU) | Standard chemotherapeutic control for comparative efficacy studies. | Typically used in micromolar range (e.g., 5-20 μM); resistance models require long-term stepwise adaptation [77]. |
| Cell Line Model | In vitro system for mechanistic studies. | MCF-7 cells: ERα-positive, wild-type p53, expresses functional A1R [4]. |
| MTT Assay Kit | Colorimetric measurement of cell viability and metabolic activity. | Standard kit for high-throughput screening of drug toxicity and proliferation [4]. |
| Annexin V-FITC Apoptosis Kit | Flow cytometry-based detection and quantification of apoptotic cells. | Distinguishes between early and late apoptosis (Annexin V+/PI- and Annexin V+/PI+) [4]. |
| qRT-PCR Reagents | Quantitative analysis of gene expression changes in apoptotic pathways. | Primers for p53, Caspase-3, -8, -9; requires RNA extraction and cDNA synthesis kits [4]. |
| siRNA for A1R | Genetic knockdown to validate pharmacological findings and study A1R function. | Validates that observed effects are specifically due to A1R loss-of-function [6]. |
This comparative analysis positions A1R antagonism as a mechanistically distinct and potent strategy worthy of further investigation in breast cancer therapeutics. The evidence suggests that targeting the A1 receptor with agents like DPCPX can effectively induce apoptosis and reduce cell viability in model systems, potentially circumventing some of the common resistance pathways that limit the efficacy of 5-FU. The cross-talk between A1R and ERα signaling presents a particularly intriguing avenue for research, especially in hormone receptor-positive breast cancers.
Future research should focus on several key areas:
- Expanding Model Systems: Investigating the effect of A1R antagonists in a broader panel of breast cancer cell lines, including those that are ER-negative, HER2-positive, and triple-negative, as well as models with acquired resistance to 5-FU and other chemotherapies.
- *In Vivo Validation: Confirming the efficacy and safety of A1R antagonists in preclinical animal models of breast cancer.
- Combination Therapy: Exploring potential synergistic effects of combining A1R antagonists with standard chemotherapeutics like 5-FU or with endocrine therapies, which may allow for lower doses of cytotoxic drugs and reduced side effects.
The preliminary data provides a compelling foundation for the continued exploration of the adenosine A1 receptor as a novel therapeutic target in the ongoing fight against breast cancer.
Validation of A1R as a Shared Antitumor Target through Multi-Compound Screening
The adenosine A1 receptor (A1R) has emerged as a critical and shared therapeutic target in breast cancer, validated through an integrated approach of multi-compound screening, computational biology, and experimental pharmacology. This technical guide details the methodology and findings from a study that identified A1R as a common target of structurally diverse antitumor compounds. By employing pharmacophore modeling, molecular docking, dynamics simulations, and in vitro assays, researchers demonstrated that A1R mediates potent antiproliferative effects in breast cancer cells. The rationally designed molecule originating from this workflow exhibited significant antitumor activity against MCF-7 cells (IC₅₀ = 0.032 µM), substantially outperforming the positive control 5-FU (IC₅₀ = 0.45 µM). These findings position A1R as a promising target for future breast cancer therapeutics and provide a validated framework for target identification and validation in oncology drug discovery.
Within the context of breast cancer research, the adenosine A1 receptor (A1R), encoded by the ADORA1 gene, represents a G-protein-coupled receptor (GPCR) with significant yet underexplored therapeutic potential. Clinical evidence indicates that A1R is ubiquitously expressed in breast cancers, providing prognostic implications for diagnosis and treatment [79]. Beyond mere presence, A1R functions within a critical feed-forward loop with estrogen receptor-α (ERα) signaling, a principal pathway in hormone-responsive breast cancer. Research has established that estradiol (E2) upregulates A1R mRNA and protein levels in ERα-positive breast cancer cells, an effect reversible by the ER antagonist ICI 182,780 [26]. Intriguingly, A1R ablation reduces ERα levels and impairs its transcriptional activity, indicating that A1R is both regulated by and necessary for full ERα function [26]. This reciprocal relationship positions A1R as a key node in hormonal signaling networks that drive breast cancer proliferation, making it a compelling target for therapeutic intervention.
Computational Workflow for Target Identification and Validation
Initial Compound Screening and Pharmacophore Modeling
The identification of A1R began with the systematic analysis of 23 compounds from published literature, each demonstrating significant inhibitory effects on MDA-MB and MCF-7 breast cancer cell lines [9] [19]. Researchers performed three-dimensional quantitative structure-activity relationship (3D-QSAR) analyses to evaluate spatial diversity, generating 249 distinct conformers through conformational optimization. Split analysis constructed five pharmacophore models, from which the most potent compound from each category was selected for further investigation (Table 1) [9] [19].
Table 1: Selected Potent Antitumor Compounds for Target Screening
| Compound | Structural Features | IC₅₀ MCF-7 (µM) | IC₅₀ MDA-MB (µM) | Citation |
|---|---|---|---|---|
| 1 | Not specified | 3.4 | 4.7 | [19] |
| 2 | Not specified | 0.21 | 0.16 | [19] |
| 3 | Not specified | 3.0 | 2.5 | [19] |
| 4 | Not specified | 0.57 | 0.42 | [19] |
| 5 | Not specified | 3.47 | 1.43 | [19] |
The SwissTargetPrediction database was employed to predict potential protein targets for these five structurally diverse compounds, using their chemical structures as input with "Homo sapiens" specified as the species [9] [19]. An intersection analysis of the predicted targets revealed A1R as a shared target across multiple compounds, highlighting its potential broad relevance in breast cancer treatment [9] [19].
Molecular Docking and Binding Affinity Assessment
Molecular docking simulations were performed to evaluate binding interactions between the selected compounds and the human adenosine A1 receptor-Gi2 protein complex (PDB ID: 7LD3) [9] [19]. Researchers created a ligand library using Discovery Studio 2019 Client and performed docking with CHARMM to refine ligand shapes and charge distribution [9]. The LibDock scores from docking simulations provided quantitative measures of binding affinity, with scores above 130 considered significant [9] [19].
Table 2: Molecular Docking Scores (LibDock) for Compounds with A1R (7LD3)
| Compound | LibDock Score | Absolute Energy | Relative Energy |
|---|---|---|---|
| 1 | 102.325 | 66.3654 | 9.57802 |
| 2 | 116.588 | 39.6037 | 1.85097 |
| 3 | 63.8847 | 56.3795 | 4.53461 |
| 4 | 130.194 | 78.0161 | 0.33005 |
| 5 | 148.673 | 53.5358 | 3.33969 |
Compound 5 exhibited the highest LibDock score (148.673) with the A1R target, suggesting superior binding affinity and making it a prime candidate for further investigation [19]. The docking poses revealed critical interaction points between the ligands and the receptor binding pocket, informing subsequent pharmacophore model development [9].
Experimental Validation of A1R as Therapeutic Target
The functional role of A1R in breast cancer proliferation was confirmed through multiple experimental approaches. RNA interference-mediated ablation of A1R in ERα-positive cells significantly reduced both basal and E2-dependent proliferation [26]. Conversely, A1R overexpression in an ERα-negative cell line induced proliferation, establishing its direct role in growth signaling [26]. Pharmacological inhibition using the selective A1R antagonist DPCPX similarly reduced proliferation, confirming A1R as a mediator of E2/ERα-dependent breast cancer growth [26].
These findings are consistent with the known signaling mechanisms of adenosine receptors, which classically modulate intracellular cAMP levels through inhibition or stimulation of adenylyl cyclase, but also activate other pathways including phospholipase C, Ca²⁺, and mitogen-activated protein kinases [80]. In breast cancer specifically, A1R appears to be required for full transcriptional activity of ERα, as A1R ablation decreases ERα binding to promoter regions of target genes like TFF1, reducing both promoter activity and mRNA levels [26].
Pathway Diagram: A1R Signaling in Breast Cancer
Diagram 1: A1R-ERα Signaling Loop in Breast Cancer. Estradiol binding to ERα upregulates A1R expression. A1R signaling modulates cAMP levels and enhances ERα transcriptional activity, creating a feed-forward loop that promotes cancer cell proliferation [26] [80].
Molecular Dynamics Simulation Protocol
System Preparation and Simulation Parameters
The stability of docked complexes between Compound 5 and A1R was evaluated using molecular dynamics (MD) simulations in GROMACS 2020.3 [9]. Protein structures were optimized with the AMBER99SB-ILDN force field, while water molecules were modeled with the TIP3P model [9]. Ligand charges were calculated using ACPYPE, which generated files compatible with the GAFF force field [9].
Simulation systems employed cubic boxes with a minimum atom-box boundary distance of 0.8 nm, hydrated with SOL water at 1000 g/L density [9]. Electrical neutrality was maintained by replacing solvent water molecules with chloride ions. The simulation protocol followed these steps:
- Energy Minimization: An initial energy minimization step relaxed the system to remove steric clashes and unfavorable interactions.
- Equilibration: A 150 ps restrained MD simulation was performed at 298.15 K to equilibrate the system while preserving protein structure.
- Production Run: Unrestricted MD simulations with a time step of 0.002 ps were performed for 15 ns, maintaining isothermal-isobaric conditions at 298.15 K and 1 bar pressure controlled by thermostats and barostats [9].
Trajectory Analysis and Binding Stability Assessment
The motion trajectory of Compound 5 interacting with A1R was analyzed using VMD 1.9.3 software [9]. Data was captured every 200 frames from the initial to the 8220th frame, allowing meticulous observation of molecular dynamics throughout the simulation timeframe [9]. This systematic approach elucidated the compound's dynamic behavior during binding, potential intermediate states, and temporal evolution of protein-ligand interactions, confirming the stability of the Compound 5-A1R complex [9].
Experimental Workflow Diagram
Diagram 2: A1R Validation Workflow. Integrated computational and experimental approach for target identification and validation, culminating in rational design of novel therapeutic candidates [9] [19].
Research Reagent Solutions Toolkit
Table 3: Essential Research Reagents for A1R-Targeted Breast Cancer Research
| Reagent/Resource | Specifications | Application | Function |
|---|---|---|---|
| A1R Antagonist | DPCPX (Selective) | Functional Studies | Pharmacological inhibition of A1R to confirm target involvement in proliferation [26] |
| siRNA/shRNA | Human ADORA1-targeting | Genetic Knockdown | RNA interference-mediated A1R ablation to assess effects on proliferation and ERα signaling [26] |
| Cell Lines | MCF-7 (ER+), MDA-MB (ER-) | In Vitro Models | Representative models of different breast cancer subtypes for efficacy testing [9] [19] |
| Docking Software | Discovery Studio 2019 | Computational Screening | Molecular docking simulations with CHARMM force field for binding affinity assessment [9] |
| MD Simulation | GROMACS 2020.3 | Dynamics Analysis | Evaluation of protein-ligand complex stability using AMBER99SB-ILDN force field [9] |
| A1R Structure | PDB ID: 7LD3 | Structural Biology | Human adenosine A1 receptor-Gi2 protein complex for structure-based drug design [9] [19] |
| Target Prediction | SwissTargetPrediction | Bioinformatics | Prediction of potential protein targets for compounds based on structural similarity [9] [19] |
The multi-compound screening approach detailed in this guide successfully validated A1R as a shared antitumor target in breast cancer. The integrated methodology combining computational predictions with experimental validation provides a robust framework for target identification and credentialing in oncology drug discovery. The finding that A1R participates in a feed-forward loop with ERα signaling not only explains its therapeutic relevance but also suggests potential for targeting A1R in hormone-responsive breast cancers [26].
The computational workflow enabled the rational design of novel therapeutic candidates, with one resulting molecule (Molecule 10) demonstrating exceptional potency (IC₅₀ = 0.032 µM) in MCF-7 cell assays [9]. This represents approximately 14-fold greater potency than the positive control 5-FU, highlighting the potential of structure-based drug design when applied to validated targets [9].
These findings position A1R as a promising therapeutic target in breast cancer and provide a comprehensive methodological framework that can be adapted for target validation campaigns against other emerging oncology targets. The convergence of computational predictions with experimental confirmation across multiple compound classes strengthens the evidence for A1R's fundamental role in breast cancer pathobiology and its therapeutic relevance.
Contrasting the Roles of A1R with Other Adenosine Receptors (A2A, A2B) in Breast Cancer
Adenosine, a purine nucleoside, is a key signaling molecule that accumulates in the hypoxic tumor microenvironment (TME) of breast cancer and other malignancies [73]. It functions as an autacoid, producing localized effects by activating a family of four G-protein coupled receptors (GPCRs): A1, A2A, A2B, and A3 [81]. These adenosine receptors (ARs) exhibit distinct expression patterns, adenosine affinities, and downstream signaling pathways, allowing them to differentially influence breast cancer pathogenesis. The current scientific literature reveals a significant research focus on the A2A and A2B receptor subtypes in breast cancer, while the role of A1R remains less characterized, representing a critical gap and opportunity for preliminary investigation [81] [73]. This whitepaper systematically contrasts the known functions of A1R with A2A and A2B receptors in breast cancer, providing a foundational context for future research into the under-explored A1R subtype.
Table 1: Fundamental Properties of Adenosine Receptors in Breast Cancer
| Receptor | Adenosine Affinity | G-Protein Coupling | Primary Signaling Pathway | Expression in Breast TME |
|---|---|---|---|---|
| A1R | High (nM range) | Gᵢ/o | ↓ cAMP, ↑ Ca²⁺ | Less defined; requires further investigation |
| A2AR | High (nM range) | Gₛ | ↑ cAMP, PKA activation | Immune cells (T cells, Myeloid cells), some cancer cells |
| A2BR | Low (μM range) | Gₛ, Gq | ↑ cAMP, ↑ Ca²⁺, MAPK modulation | Breast cancer cells (esp. TNBC), endothelial cells, immune cells |
Receptor Expression and Signaling Pathways
The differential effects of adenosine receptors in breast cancer are fundamentally driven by their unique signaling mechanisms. The high-affinity A2AR and low-affinity A2BR have been the subject of extensive mapping due to their pronounced roles in immune suppression and cancer stemness.
A2A and A2B Receptor Signaling in Breast Cancer
The A2B receptor (A2BR) has emerged as a particularly significant player in aggressive breast cancer subtypes. In triple-negative breast cancer (TNBC) cells, high A2BR expression is associated with the enrichment of breast cancer stem cells (BCSCs), a subpopulation responsible for tumor initiation, metastasis, and therapy resistance [82] [83]. Research demonstrates that chemotherapy (e.g., paclitaxel, carboplatin) induces A2BR protein expression, which activates a complex signaling cascade promoting BCSC specification. This pathway involves A2BR-mediated activation of p38 MAPK, leading to the nuclear translocation of the chromatin remodeling factor SMARCD3. SMARCD3 then recruits histone-modifying enzymes (KDM6A and p300) to the regulatory regions of pluripotency factor genes NANOG, SOX2, and KLF4. This recruitment decreases repressive H3K27me3 marks and increases activating H3K27ac marks, facilitating transcription factor FOXO3 binding and ultimately driving the transcription of these core pluripotency factors [82] [83]. In contrast, studies in MDA-MB-231 cells show that A2BR activation can also reduce phosphorylation of ERK1/2, a key MAPK, through a mechanism involving cAMP/PKA and intracellular Ca²⁺, which stimulates MAPK phosphatase-1 (MKP-1) activity. This context-dependent suppression of MAPK signaling may represent a novel growth-regulatory mechanism [84].
The high-affinity A2AR is primarily recognized for its potent immunosuppressive effects within the TME. Upon activation by adenosine, it triggers a Gₛ-mediated increase in intracellular cAMP in various immune cells, including T cells and myeloid cells, leading to their functional inactivation and fostering an environment conducive to tumor immune evasion [73].
A1 Receptor Signaling: A Knowledge Gap
The signaling role of A1R in breast cancer cells remains poorly defined. Based on its canonical signaling in other tissues, A1R is known to couple to Gᵢ/o proteins, leading to the inhibition of adenylyl cyclase and a reduction in intracellular cAMP levels. It can also activate phospholipase C (PLC) via Gᵢ/o-derived βγ subunits, resulting in increased intracellular Ca²⁺ mobilization [81]. However, the specific downstream targets, functional consequences, and potential cross-talk of this pathway within the context of breast cancer biology are areas ripe for preliminary investigation. The contrasting signaling outputs of A1R (↓cAMP) versus A2AR/A2BR (↑cAMP) suggest potentially opposing roles in regulating cancer cell proliferation, survival, and communication with the TME.
Diagram 1: Adenosine receptor signaling pathways in breast cancer. A1R signaling remains less defined, while A2A/A2BR signaling influences cAMP, MAPK pathways, and pluripotency.
Functional Roles in Breast Cancer Pathogenesis
The distinct signaling profiles of A2A and A2B receptors translate into specific and critical roles in breast cancer progression, metastasis, and therapy resistance. The functional impact of A1R activation, however, is not well-documented in the available literature.
Documented Roles of A2A and A2B Receptors
The A2B receptor serves as a key mediator of therapy-induced resistance and stemness. In TNBC, chemotherapy-induced A2BR expression directly promotes the enrichment of BCSCs, a primary driver of tumor recurrence and metastasis [82] [83]. Genetic knockdown or pharmacological inhibition of A2BR (e.g., with alloxazine) blocks chemotherapy-induced expression of pluripotency factors and BCSC enrichment, both in vitro and in vivo, and delays tumor recurrence after chemotherapy is discontinued [83]. This positions A2BR as a high-value therapeutic target for combination therapy in aggressive breast cancers.
Both A2A and A2B receptors are potent contributors to an immunosuppressive TME. Activation of these receptors on immune cells—including monocytes, dendritic cells, and T cells—promotes an immunosuppressive phenotype. Notably, β2-AR activation (from adrenergic signaling) is associated with high PD-L1 expression on BC cells, while β-AR blockade downregulates PD-1 on T cells and increases interferon-γ production [85]. This immunomodulatory role provides a strong rationale for combining AR antagonists with immunotherapy. Furthermore, A2BR activation on breast cancer cells stimulates the production of vascular endothelial growth factor (VEGF), thereby promoting angiogenesis and tumor growth [85].
Contrasting and Potential Role of A1R
In stark contrast to A2A and A2B receptors, the role of A1R in breast cancer pathogenesis is not defined in the current search results. A review article from 2025 notes that among P1 receptors, "A1, A2A, and A2B receptors are involved in the proliferation and invasion of breast cancer, while the A3 receptor is related to the inhibition of tumor growth" [81]. This statement suggests a potential tumor-promoting role for A1R, but specific mechanistic studies and functional data are lacking. This significant knowledge gap highlights the critical need for preliminary investigations to determine whether A1R activity promotes proliferation, invasion, and metastasis akin to A2BR, or whether it exerts contrasting, potentially protective effects.
Table 2: Functional Roles of Adenosine Receptors in Breast Cancer Pathogenesis
| Receptor | Role in Cancer Stemness | Role in Immune Modulation | Role in Angiogenesis | Response to Therapy |
|---|---|---|---|---|
| A1R | Unknown | Unknown | Unknown | Unknown |
| A2AR | Not a primary role | Potent immunosuppression; inhibits T cell function and promotes exhaustion | Minor role | Potential target to boost immunotherapy |
| A2BR | Critical: Promotes BCSC enrichment and pluripotency factor expression via epigenetic regulation | Contributes to immunosuppression; associated with PD-L1 expression | Strong: Stimulates VEGF production | Induces chemoresistance; target to prevent relapse |
Experimental and Therapeutic Landscape
The disparate roles of adenosine receptors are reflected in the focused development of targeted therapies for A2A and A2B receptors, while A1R remains largely unexplored as a drug target in oncology.
Targeted Agents and Clinical Translation
The therapeutic inhibition of the adenosine pathway is a rapidly advancing area in oncology, with a clear emphasis on A2A and A2B receptors. Multiple selective antagonists are in clinical development. For A2BR, PBF-1129 is in Phase I trials for lung cancer, and TT-4 is in Phase I/II trials for advanced solid tumors [81]. The safety profile of AR antagonists is considered favorable, bolstered by the approval of the A2AR antagonist istradefylline for Parkinson's disease [81]. Preclinical data strongly supports the use of A2BR antagonists in breast cancer. Combining A2BR inhibition with chemotherapy has been shown to block BCSC enrichment and significantly delay tumor recurrence in mouse models of TNBC, presenting a promising strategy to overcome chemoresistance [82] [83].
Research Reagents and Methodologies
Investigating the adenosine pathway in breast cancer requires a specialized toolkit of reagents and validated experimental protocols. The following table details key resources for probing receptor function and its biological consequences.
Table 3: Research Reagent Solutions for Adenosine Receptor Studies
| Reagent / Assay | Function / Target | Example Specific Agents | Key Application in Breast Cancer Research |
|---|---|---|---|
| A2BR Antagonists | Inhibits A2BR signaling | Alloxazine, PBF-1129 | Block chemotherapy-induced BCSC enrichment; delay tumor recurrence in vivo [83] |
| A2BR Agonists | Activates A2BR signaling | NECA (non-selective) | Study receptor signaling outcomes (e.g., ERK1/2 dephosphorylation, MKP-1 activation) [84] |
| Aldefluor Assay | Measures ALDH enzyme activity | ALDH substrate (BAAA) | Identify and quantify breast cancer stem cell (BCSC) population in vitro [82] [83] |
| Mammosphere Assay | Assesses self-renewal capacity | Ultra-low attachment plates | Evaluate stemness and tumor-initiating potential of cells in vitro [82] [83] |
| ChIP Assay | Maps protein-DNA interactions | Antibodies against H3K27ac, H3K27me3, FOXO3 | Investigate epigenetic regulation of pluripotency factors (NANOG, SOX2, KLF4) [82] |
| shRNA Knockdown | Genetically depletes target gene | shRNA targeting ADORA2B | Validate specific role of A2BR in BCSC phenotype and therapy resistance [83] |
To systematically evaluate the role of adenosine receptors, particularly in stemness and therapy resistance, the following integrated experimental workflow is employed in preclinical studies.
Diagram 2: Experimental workflow for studying adenosine receptors in breast cancer stemness.
The roles of adenosine receptors in breast cancer are profoundly contrasting. The A2A receptor is established as a master regulator of immunosuppression, while the A2B receptor is a multi-faceted oncoprotein driving cancer stemness, angiogenesis, and chemoresistance in aggressive subtypes like TNBC. This functional understanding has propelled the development of A2A and A2B antagonists into the clinical trial pipeline. In sharp focus, however, is the role of the A1 receptor, which remains virtually unexplored territory within breast cancer research. The preliminary statement that A1R may be involved in proliferation and invasion is a compelling hypothesis that lacks mechanistic depth. Future preliminary investigations must prioritize defining A1R's expression patterns across breast cancer molecular subtypes, its impact on core hallmarks of cancer (proliferation, apoptosis, invasion), and its potential cross-talk with the well-characterized A2B pathway. Elucidating the function of A1R could unveil novel biological insights and unexpected therapeutic opportunities, potentially completing the complex picture of adenosine-mediated signaling in breast cancer.
The preliminary investigation of the adenosine A1 receptor (A1R) in breast cancer research reveals a complex and promising therapeutic target. Adenosine receptors are a family of G protein-coupled receptors comprising four subtypes (A1, A2A, A2B, and A3), with the A1 receptor demonstrating dual functionality in cancer pathophysiology. While this receptor can mediate proliferative effects in certain contexts, targeted pharmacological intervention demonstrates significant antitumor potential, positioning A1R as a compelling subject for preclinical drug development. This whitepaper synthesizes current preclinical evidence, highlighting the critical quantitative metrics and methodological approaches that establish A1R's role in oncology. The compelling data from in vitro cytotoxicity assays and in vivo xenograft models provide a robust foundation for framing A1R within a broader thesis of innovative cancer therapeutics, offering researchers a detailed roadmap for future investigation.
The Adenosine A1 Receptor in Breast Cancer: A Dual-Faced Target
The adenosine A1 receptor exhibits a paradoxical role in breast cancer biology, functioning both as a promoter of proliferation and a potential therapeutic target. Research indicates that A1R is an E2/ERα target gene and a regulator of ERα transcriptional activity, forming a short feed-forward loop that favors breast cancer growth. In ERα-positive breast cancer cells, estradiol (E2) upregulates A1R mRNA and protein levels, an effect reversible by the E2 antagonist ICI 182,780 [6]. This establishes A1R as a direct mediator of hormone-dependent cancer progression. Consequently, A1R ablation or pharmacological antagonism reduces basal and E2-dependent proliferation, establishing its critical role in cancer growth [6].
However, this very proliferative mechanism can be therapeutically exploited. The A1 receptor antagonist 1,3-dipropyl-8-cyclopentylxanthine (DPCPX) significantly induces apoptosis in MCF-7 cells, reduces cell viability, and dramatically up-regulates the expression of p53 and caspases 3, 8, and 9 [5]. This demonstrates that targeted inhibition of A1R can activate pro-apoptotic signaling pathways, effectively switching the receptor's function from pro-survival to pro-death. This dual nature underscores the importance of context and therapeutic strategy when investigating A1R in oncology, making it a compelling target for drug development.
Quantitative Analysis of Antitumor Efficacy
In Vitro Cytotoxicity and IC50 Values
The half-maximal inhibitory concentration (IC50) serves as a primary quantitative metric for evaluating the potency of therapeutic compounds in preclinical research. The following table summarizes key IC50 values for various compounds targeting adenosine receptors and other novel targets in breast cancer models.
Table 1: In Vitro Cytotoxicity (IC50) of Selected Compounds in Breast Cancer Models
| Compound | Target/Cell Line | IC50 Value | Biological Effect | Source/Reference |
|---|---|---|---|---|
| Molecule 10 | MCF-7 Breast Cancer Cells | 0.032 µM | Antitumor activity | [9] |
| 5-FU (Control) | MCF-7 Breast Cancer Cells | 0.45 µM | Antitumor activity (positive control) | [9] |
| DPCPX (A1R Antagonist) | MCF-7 Cells (72h treatment) | Significant reduction in viability (qualitative) | Induced apoptosis, upregulated p53 and caspases | [5] |
| Imidazothioxanthone 10a | NCI 60 Human Tumor Cell Line Panel | 6.8 µM | General cytotoxic activity | [86] |
| Imidazothioxanthone 10c | NCI 60 Human Tumor Cell Line Panel | 8.3 µM | General cytotoxic activity | [86] |
The data demonstrates a striking potency of Molecule 10, a compound designed based on adenosine A1 receptor targeting, showing a significantly lower IC50 (0.032 µM) than the positive control 5-FU (0.45 µM) in MCF-7 cells [9]. This indicates superior cytotoxic potency in vitro. Furthermore, the efficacy of the A1R antagonist DPCPX, while not quantified with a specific IC50, is clearly evidenced by its significant induction of apoptosis and upregulation of key tumor suppressor and executioner proteins [5].
In Vivo Antitumor Activity in Xenograft Models
In vivo validation is a critical step in the preclinical drug development pipeline, providing essential data on compound efficacy in a complex living system. The following table collates results from key in vivo studies using human and murine tumor models.
Table 2: In Vivo Antitumor Efficacy in Preclinical Xenograft Models
| Compound | In Vivo Model | Dosing & Regimen | Key Efficacy Results | Source/Reference |
|---|---|---|---|---|
| Imidazothioxanthone 10a | Human Mammary Carcinoma MT-1 | Not specified | Active against tumor growth | [86] |
| Murine Colon Cancer MAC15A | Not specified | Active against tumor growth | [86] | |
| Human Ovarian Cancer PXN/109T/C | Not specified | Marginal activity | [86] | |
| Human Mammary Cancer MCF-7 | Not specified | Inactive | [86] | |
| BYS10 (RET inhibitor) | Ba/F3-KIF5B-RET Xenograft | 3 mg/kg, orally, twice daily | TGI: 78.45% (vs. 57.06% for Selpercatinib) | [87] |
| Ba/F3-KIF5B-RET-V804L Xenograft | 3 mg/kg, orally, twice daily | TGI: 94.67% (vs. 79.48% for Selpercatinib) | [87] |
The in vivo data for compound 10a reveals a model-dependent antitumor effect, showing clear activity in some solid tumor models (MT-1, MAC15A) while being inactive in others (MCF-7) [86]. This underscores the necessity of testing candidate compounds across a diverse panel of relevant models. For context, the table also includes data on BYS10, a novel RET inhibitor, which demonstrates the high level of antitumor efficacy (Tumor Growth Inhibition, TGI) that can be achieved with targeted therapies, providing a benchmark for potency in the field [87].
Experimental Protocols for Key Assays
In Vitro Cell Viability and Cytotoxicity Assays
Objective: To quantitatively determine the effect of adenosine A1 receptor ligands on breast cancer cell proliferation and viability. Principle: This assay measures the conversion of a substrate by metabolically active cells, serving as a proxy for cell viability and proliferation. Key Reagents: Sulforhodamine B (SRB) assay, MTT assay, or CellTiter-Glo luminescent assay. Detailed Procedure:
- Cell Seeding: Plate MCF-7 or other breast cancer cell lines in 96-well plates at a density of 5,000-10,000 cells per well in complete medium and culture for 24 hours.
- Compound Treatment: Serially dilute the test compounds (e.g., A1R agonist CPA, antagonist DPCPX, or novel compounds like Molecule 10). Replace the medium with fresh medium containing the compounds or vehicle control (DMSO). Incubate for a predetermined time (e.g., 72 hours).
- Viability Measurement:
- For MTT: Add MTT reagent (0.5 mg/mL final concentration) and incubate for 2-4 hours. Solubilize the formed formazan crystals with DMSO and measure the absorbance at 570 nm.
- For SRB: Fix cells with trichloroacetic acid, stain with SRB, and dissolve the bound dye for absorbance reading.
- For CellTiter-Glo: Add an equal volume of CellTiter-Glo reagent to lyse cells and generate a luminescent signal proportional to ATP content.
- Data Analysis: Calculate the percentage of viability relative to the vehicle control. Use non-linear regression analysis to determine the IC50 values from dose-response curves [86] [9] [5].
Molecular Docking and Dynamics Simulations
Objective: To predict and analyze the binding mode and stability of a candidate compound (e.g., Molecule 10) within the adenosine A1 receptor binding pocket. Principle: Computational methods simulate the interaction between a small molecule (ligand) and a protein target, providing insights into binding affinity and mechanism at the atomic level. Key Reagents: Protein Data Bank structure of the target (e.g., A1R-Gi protein complex, PDB ID: 7LD3), docking software (e.g., AutoDock Vina), molecular dynamics software (e.g., GROMACS). Detailed Procedure:
- System Preparation:
- Obtain the 3D structure of the human adenosine A1 receptor from the PDB.
- Prepare the ligand structure by energy minimization and assign appropriate charges.
- Molecular Docking:
- Define the binding site coordinates, often based on the known location of co-crystallized ligands.
- Perform flexible or rigid docking simulations to generate multiple potential binding poses.
- Score the poses based on binding energy and select the most favorable pose for further analysis [9].
- Molecular Dynamics (MD) Simulation:
- Embed the docked protein-ligand complex in a solvated lipid bilayer to mimic the cell membrane.
- Assign force field parameters (e.g., AMBER99SB-ILDN for the protein, GAFF for the ligand).
- Perform an energy minimization to remove steric clashes.
- Equilibrate the system with restrained heavy atoms, followed by unrestricted production MD runs (e.g., 15-100 ns) under isothermal-isobaric conditions (e.g., 298.15 K, 1 bar).
- Trajectory Analysis: Analyze the root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), and ligand-protein interaction profiles over the simulation time to assess complex stability [9].
In Vivo Efficacy Study in Xenograft Models
Objective: To evaluate the antitumor efficacy of an A1R-targeting compound in a live animal model of breast cancer. Principle: Human cancer cells are implanted into immunodeficient mice to form tumors, allowing for the assessment of tumor growth inhibition in response to drug treatment. Key Reagents: Breast cancer cells (e.g., MCF-7, MT-1), immunodeficient mice (e.g., NOD SCID, B-NDG), test and control compounds. Detailed Procedure:
- Tumor Inoculation: Subcutaneously inject a suspension of breast cancer cells (e.g., 1x10^6 MCF-7 cells resuspended in Matrigel) into the flank of female immunodeficient mice.
- Randomization and Dosing: When the average tumor volume reaches a predetermined size (e.g., 80-150 mm³), randomly assign mice into treatment and control groups (n=6-8). Commence dosing via the chosen route (e.g., oral gavage, intraperitoneal injection). The control group receives the vehicle.
- Monitoring and Endpoint Measurement:
- Monitor body weight and general health regularly.
- Measure tumor dimensions 2-3 times per week using a caliper. Calculate tumor volume using the formula: Volume = (Length × Width²) / 2.
- Continue treatment for a set duration (e.g., 3-4 weeks) or until tumor burden endpoints are reached.
- Data Analysis and Reporting:
- Plot mean tumor volume ± SEM for each group over time.
- Calculate the percent tumor growth inhibition (TGI) at the end of the study: TGI (%) = [1 - (Tumor Volumetreated / Tumor Volumecontrol)] × 100%.
- Perform statistical analysis (e.g., one-way ANOVA) to determine significance [86].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagent Solutions for Investigating A1R in Cancer
| Reagent/Category | Specific Examples | Function & Application |
|---|---|---|
| A1R Agonists | N⁶-Cyclopentyladenosine (CPA) | Activates A1R signaling; used to study receptor activation effects on cell proliferation, viability, and pathway analysis. |
| A1R Antagonists | 1,3-dipropyl-8-cyclopentylxanthine (DPCPX) | Inhibits A1R signaling; used to probe the functional consequences of A1R blockade, including apoptosis induction. |
| Cell Lines | MCF-7 (ER⁺), MDA-MB-231 (ER⁻) | Model systems for in vitro experiments (viability, apoptosis, signaling) and in vivo xenograft generation. |
| Antibodies | Anti-p53, Anti-Cleaved Caspase-3/8/9, Anti-phospho-ERK, Anti-ERα | Detect protein expression and activation (by Western blot, immunofluorescence) in mechanistic studies. |
| Computational Tools | AutoDock Vina, GROMACS, PDB Structure 7LD3 | Molecular modeling, docking, and dynamics simulations to study ligand-receptor interactions and stability. |
Signaling Pathways and Experimental Workflows
A1 Receptor Signaling and Therapeutic Intervention in Breast Cancer
Diagram 1: A1 Receptor Signaling and Therapeutic Intervention in Breast Cancer. This diagram illustrates the dual pathways: A1R agonism promotes proliferation via Gi-mediated inhibition of adenylyl cyclase, reduced cAMP, and modulation of ERα activity. In contrast, A1R antagonism (e.g., DPCPX) induces apoptosis via p53 and caspase upregulation [5] [6] [23].
Preclinical Drug Discovery Workflow
Diagram 2: Preclinical Drug Discovery Workflow. This flowchart outlines the key stages in the preclinical development of A1R-targeting therapeutics, from initial target identification and compound design to in vitro and in vivo validation, forming an iterative optimization cycle [86] [9].
The collective preclinical evidence firmly positions the adenosine A1 receptor as a therapeutically relevant target in breast cancer. The quantitative data, particularly the low IC50 values of novel compounds and the efficacy demonstrated in vivo, provides a compelling argument for continued investment in this research area. The methodological frameworks for cytotoxicity assessment, mechanistic investigation, and in vivo validation, as detailed in this whitepaper, offer a robust and reproducible pathway for researchers to evaluate new A1R-targeting entities. While the receptor's biology is complex, the strategic application of antagonists or allosteric modulators can effectively harness this system to induce pro-apoptotic and anti-proliferative outcomes. Future work should focus on optimizing the selectivity and pharmacokinetic properties of lead compounds, exploring combination therapies, and further elucidating the precise molecular mechanisms by which A1R modulation exerts its potent antitumor effects, as illustrated in the provided signaling pathway.
The adenosinergic pathway has emerged as a critical immunosuppressive mechanism within the tumor microenvironment across multiple cancer types. This review synthesizes pan-cancer genomic and epigenetic analyses of adenosine pathway components, with specific emphasis on their implications for breast cancer pathogenesis and the preliminary investigation of adenosine A1 receptor (ADORA1) as a therapeutic target. Accumulating evidence reveals significant heterogeneity in the alteration patterns of adenosine-generating enzymes and receptors across cancer types, with breast cancer demonstrating distinct expression profiles linked to hormone receptor status and metastatic behavior. Integration of computational, biochemical, and functional data suggests that targeted inhibition of specific adenosine pathway components, particularly ADORA1, represents a promising strategy for overcoming immunosuppression and enhancing anti-tumor immunity in breast cancer.
Extracellular adenosine accumulation represents a fundamental immunosuppressive mechanism exploited by cancers to evade host immune surveillance. Under hypoxic conditions characteristic of solid tumors, ATP is rapidly released into the extracellular space where it undergoes sequential hydrolysis to adenosine through the coordinated action of ectonucleotidases [88]. The CD39-CD73 axis constitutes the primary pathway for extracellular adenosine production, with CD39 (ENTPD1) catalyzing the conversion of ATP/ADP to AMP, and CD73 subsequently dephosphorylating AMP to adenosine [88]. Alternative, non-canonical pathways involving CD38 and CD203a also contribute to adenosine generation, particularly under hypoxic conditions [88].
Once generated, extracellular adenosine exerts its effects primarily through binding to four G-protein-coupled P1 purinergic receptors (A1, A2A, A2B, and A3), which exhibit differing affinities for adenosine and downstream signaling consequences [88]. The immunosuppressive properties of adenosine are primarily mediated through A2A and A2B receptors, leading to increased intracellular cAMP levels in immune cells and subsequent inhibition of effector functions [88]. The adenosine pathway thus represents a promising therapeutic target for cancer immunotherapy, with multiple inhibitory strategies currently under investigation.
Pan-Cancer Genomic and Epigenetic Landscape of Adenosine Pathway Components
Genomic Alterations Across Cancer Types
Comprehensive pan-cancer analysis reveals significant heterogeneity in the genomic and epigenetic profiles of adenosinergic pathway components. Mutation frequencies of adenosine pathway genes are generally low across most cancer types, though specific patterns of dysregulation emerge [88]. The table below summarizes key alterations in major adenosine pathway components across selected cancer types:
Table 1: Pan-Cancer Genomic and Expression Alterations in Adenosine Pathway Components
| Component | Alteration Type | Cancer Types with Significant Alterations | Functional Consequences |
|---|---|---|---|
| CD73 (NT5E) | Overexpression | Multiple including breast, ovarian, pancreatic | Increased adenosine production, immune suppression, metastasis |
| CD39 (ENTPD1) | Overexpression | Breast, lung, pancreatic | Enhanced ATP-to-AMP conversion, tumor progression |
| A1 Receptor (ADORA1) | Variable expression | Breast (ER+), cholangiocarcinoma, ovarian | Context-dependent pro-or anti-tumor effects |
| A2A Receptor (ADORA2A) | Overexpression | Multiple cancer types | T-cell anergy, enhanced Treg function |
| A2B Receptor (ADORA2B) | Overexpression | Breast, colorectal, pancreatic | Angiogenesis, cytokine production |
| A3 Receptor (ADORA3) | Variable expression | Breast, melanoma, prostate | Cell motility arrest at high agonist concentrations |
| ADA1 | Overexpression | Cholangiocarcinoma, DLBCL, ovarian, pancreatic | Poor prognosis in multiple cancers |
| ADA2 | Overexpression | Esophageal, glioblastoma, acute myeloid leukemia | Favorable prognosis in breast, ovarian, other cancers |
Epigenetic Regulation in Breast Cancer
Epigenetic mechanisms, including DNA methylation, histone modifications, and non-coding RNA regulation, play crucial roles in modulating adenosine pathway components in breast cancer. In triple-negative breast cancer (TNBC), DNA methylation patterns significantly influence the expression of extracellular matrix (ECM) components and epithelial-to-mesenchymal transition (EMT) factors that interact with adenosine signaling [89]. Specifically, hypermethylation of the E-cadherin gene (CDH1) promoter collaborates with histone deacetylation by HDAC1 and HDAC2 to silence expression, facilitating EMT and potentially enhancing adenosine-mediated immunosuppression [89].
MicroRNA networks further regulate the adenosinergic pathway in breast cancer, with miR-200 family members critically influencing EMT and cancer stem cells through regulation of ZEB1, ZEB2, and BMI1 expression [89]. Loss of miR-200 expression occurs in invasive breast cancer cells and breast cancer stem cells, establishing an epigenetic link to pathways that may coordinate with adenosine signaling [89]. Additionally, enhancer of zeste homolog 2 (EZH2) can inhibit miR-200 expression to induce ZEB1 and ZEB2 expression, suggesting a critical loop between polycomb group proteins, miR-200, EMT transcription factors, and potentially adenosine pathway components [89].
Adenosine Pathway in Breast Cancer: Focus on A1 Receptor
ADORA1 as Estrogen Receptorα Regulator and Target
In the context of breast cancer, the adenosine A1 receptor (ADORA1) demonstrates a unique dual role as both a target and regulator of estrogen receptorα (ERα) action. In ERα-positive breast cancer cells, estradiol (E2) significantly upregulates ADORA1 mRNA and protein levels, an effect that is reversed by the ER antagonist ICI 182,780 [2]. This regulatory relationship establishes ADORA1 as a direct mediator of E2/ERα-dependent breast cancer growth, creating a short feed-forward loop that favors tumor proliferation.
Functional studies demonstrate that siRNA ablation of ADORA1 in ERα-positive cells reduces both basal and E2-dependent proliferation, while ADORA1 overexpression in ERα-negative cell lines induces proliferation [2]. The selective ADORA1 antagonist DPCPX similarly reduces proliferation, confirming ADORA1 as a legitimate therapeutic target in hormone-dependent breast cancer [2]. Intriguingly, ADORA1 ablation decreases mRNA and protein levels of ERα itself and consequently reduces estrogen-responsive element-dependent ERα transcriptional activity, suggesting a reciprocal regulatory relationship [2].
Receptor-Specific Effects in Breast Cancer Progression
Beyond ADORA1, other adenosine receptors demonstrate distinct functional roles in breast cancer progression. The A3 adenosine receptor (ADORA3) exhibits complex behavior in breast cancer cell motility. While autocrine adenosine signaling through ADORA3 promotes migration of MDA-MB-231 breast cancer cells, exogenous adenosine or specific A3 receptor agonists (e.g., IB-MECA) dose-dependently arrest cell motility [90]. This arrest occurs through simultaneous stimulation of multiple leading edges, doubling cell surface areas and reducing migration velocity by up to 75% [90].
The differential distribution of adenosine receptors within breast cancer cells contributes to their functional specificity. Compared to benign human mammary epithelial cells (HMEC), MDA-MB-231 cells possess multiple leading edges enriched with A3 adenosine receptors and overexpress the ectonucleotidases ENPP1 and CD73, which convert extracellular ATP to adenosine [90]. This subcellular distribution pattern of adenosine receptors contributes to dysfunctional cell motility in malignant cells.
Diagram 1: Adenosine Signaling Pathway in Breast Cancer. This diagram illustrates the generation of adenosine in the tumor microenvironment and its subsequent signaling through specific receptors, leading to functional outcomes in breast cancer cells, with emphasis on ADORA1-ERα crosstalk.
Adenosine Deaminase Isoenzymes: Distinct Roles in Breast Cancer
The adenosine deaminase isoenzymes ADA1 and ADA2, which catalyze the deamination of adenosine, demonstrate contrasting expression patterns and prognostic significance in breast cancer. A comprehensive pan-cancer analysis revealed that while both enzymes perform the same biochemical reaction, they exert distinct biological effects in the tumor microenvironment [91].
In breast invasive carcinoma (BRCA), high ADA2 expression is associated with a favorable prognosis, whereas high ADA1 expression shows opposite prognostic patterns in several other cancers [91]. ADA2 demonstrates positive correlation with B cells, CD8 T cells, monocytes/macrophages, and dendritic cells, and shows strong negative correlation with myeloid-derived suppressor cells in the breast tumor microenvironment [91]. Functional enrichment analyses further distinguish these isoenzymes, with ADA1 expression-related genes primarily involved in cell division, while ADA2-related genes are predominantly associated with immune response pathways [91].
Therapeutic Targeting and Experimental Approaches
Computational Drug Discovery for ADORA1 Targeting
Recent advances in computational approaches have facilitated the identification and optimization of compounds targeting the adenosine A1 receptor for breast cancer treatment. Integrated bioinformatics and computational chemistry strategies have identified ADORA1 as a key candidate target through intersection analysis of compounds with demonstrated efficacy against MCF-7 and MDA-MB-231 breast cancer cell lines [9] [19].
Molecular docking and dynamics simulations with the human adenosine A1 receptor-Gi2 protein complex (PDB ID: 7LD3) have enabled the evaluation of binding stability for candidate compounds [9] [19]. One such study identified Compound 5 as exhibiting stable binding to ADORA1, with a LibDockScore of 148.673, significantly outperforming other tested compounds [19]. Pharmacophore modeling based on this binding information guided the rational design of Molecule 10, which demonstrated potent antitumor activity against MCF-7 cells with an IC50 value of 0.032 µM, significantly outperforming the positive control 5-FU (IC50 = 0.45 µM) [9] [19].
Diagram 2: Experimental Workflow for ADORA1-Targeted Drug Discovery. This diagram outlines the integrated computational and experimental approach for identifying and validating adenosine A1 receptor-targeted compounds for breast cancer treatment.
Research Reagent Solutions for Adenosine Pathway Investigation
The table below outlines essential research reagents and methodologies for investigating the adenosine pathway in breast cancer research:
Table 2: Essential Research Reagents and Methodologies for Adenosine Pathway Investigation
| Reagent/Method | Specific Examples | Research Application | Key Findings Enabled |
|---|---|---|---|
| Selective Receptor Antagonists | DPCPX (A1), CSC (A2A), MRS 1754 (A2B) | Receptor-specific functional studies | ADORA1 ablation reduces ERα-positive breast cancer proliferation [2] [90] |
| Selective Receptor Agonists | IB-MECA (A3), CGS 21680 (A2A) | Receptor activation studies | A3 receptor stimulation arrests MDA-MB-231 cell motility [90] |
| siRNA/shRNA Knockdown | ADORA1-targeted siRNA | Gene function ablation | ADORA1 knockdown reduces ERα levels and transcriptional activity [2] |
| Molecular Docking | Human ADORA1-Gi2 complex (7LD3) | Compound binding analysis | Identification of Compound 5 with LibDockScore 148.673 [19] |
| Molecular Dynamics Simulations | GROMACS with AMBER99SB-ILDN force field | Protein-ligand binding stability | Validation of Compound 5-ADORA1 binding stability [9] |
| Activity Assays | Enzymatic ADA activity detection | Enzyme function measurement | Differential ADA1/ADA2 activities in cancer serum [91] |
| Cell Migration Tracking | Time-lapse video microscopy | Motility analysis | Adenosine-mediated arrest of breast cancer cell motility [90] |
The pan-cancer genomic and epigenetic landscape of the adenosine pathway reveals substantial heterogeneity in alteration patterns across cancer types, with breast cancer demonstrating unique features related to hormone receptor status and metastatic behavior. The adenosine A1 receptor emerges as a particularly promising target in breast cancer due to its dual role as both a regulator and target of ERα signaling, creating a feed-forward loop that promotes tumor growth. Preclinical evidence supports the therapeutic potential of ADORA1 targeting, with computational drug discovery approaches yielding promising candidate compounds with potent anti-tumor activity.
Future research directions should focus on validating these findings in additional breast cancer models, exploring combination therapies targeting multiple adenosine pathway components simultaneously, and developing biomarker strategies to identify patient populations most likely to benefit from adenosine-targeted therapies. The distinct roles of adenosine deaminase isoenzymes ADA1 and ADA2 in breast cancer prognosis and immune modulation further underscore the complexity of this pathway and present additional opportunities for therapeutic intervention. As our understanding of the genomic and epigenetic regulation of the adenosine pathway in breast cancer continues to evolve, so too will opportunities for translating these findings into improved clinical outcomes for breast cancer patients.
Conclusion
This preliminary investigation consolidates compelling evidence that the Adenosine A1 Receptor is a viable and promising therapeutic target in breast cancer. The integration of foundational biology with advanced computational and in vitro methodologies has enabled the rational design of potent A1R-targeting compounds, with some demonstrating superior antitumor activity compared to conventional chemotherapeutics. While challenges in receptor selectivity and on-target side effects remain, emerging strategies such as allosteric modulation offer pathways to overcome these hurdles. Future research must prioritize the translation of these preclinical findings through robust in vivo studies and, ultimately, clinical trials. Combining A1R-targeted agents with existing immunotherapies presents a particularly exciting avenue for developing novel, effective combination regimens against breast cancer.
References
- 1. Current Understanding of the Role of Adenosine Receptors in Cancer [https://www.mdpi.com/1420-3049/29/15/3501]
- 2. , a target and regulator of estrogen receptorα... Adenosine A 1 receptor [https://www.nature.com/articles/onc2009409?error=cookies_not_supported]
- 3. Adenosine Receptors and Cancer - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3598010/]
- 4. The effect of adenosine agonist and antagonist on p53... A 1 receptor [https://pmc.ncbi.nlm.nih.gov/articles/PMC5022378/]
- 5. The effect of adenosine A1 receptor agonist and antagonist on p53 and caspase 3, 8, and 9 expression and apoptosis rate in MCF-7 breast cancer cell line - PubMed [https://pubmed.ncbi.nlm.nih.gov/27651810/]
- 6. Adenosine A1 receptor, a target and regulator of estrogen receptoralpha action, mediates the proliferative effects of estradiol in breast cancer - PubMed [https://pubmed.ncbi.nlm.nih.gov/19935720/]
- 7. Frontiers | Dual Inhibition of Ornithine Decarboxylase and... [https://www.frontiersin.org/journals/oncology/articles/10.3389/fonc.2021.636373/full]
- 8. modifies P53 expression and apoptosis in... Adenosine A 1 receptor [https://pubmed.ncbi.nlm.nih.gov/27075390/]
- 9. Frontiers | Target screening and optimization of candidate compounds... [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2025.1467504/full]
- 10. Frontiers | Small molecule allosteric modulation of the adenosine... [https://www.frontiersin.org/journals/endocrinology/articles/10.3389/fendo.2023.1184360/full]
- 11. Adenosine A Receptor and Epilepsy | Encyclopedia MDPI 1 [https://encyclopedia.pub/entry/6558]
- 12. Frontiers | Editorial: Community series in adenosine pathways in... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2025.1538930/full]
- 13. The role of adenosine A receptor on immune cells | Inflammation... 1 [https://link.springer.com/article/10.1007/s00011-022-01607-w]
- 14. High PANX1 Expression Leads To Neutrophil... | Research Square [https://www.researchsquare.com/article/rs-899442/v1]
- 15. Adenosine receptor - Wikipedia [https://en.wikipedia.org/wiki/Adenosine_receptor]
- 16. Prognostic value of immune checkpoint molecules in breast cancer - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7340863/]
- 17. Rationally Engineered Adenosine 2A Decoy A for Reversing... Receptor [https://pubmed.ncbi.nlm.nih.gov/40801136/]
- 18. Immune Checkpoint Blockade for Breast Cancer - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6061922/]
- 19. Target screening and optimization of candidate compounds for breast cancer treatment using bioinformatics and computational chemistry approaches - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12098522/]
- 20. Integrated multi-omics analysis reveals the functional and prognostic... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12006012/]
- 21. Frontiers | Unlocking the adenosine mechanism of the... receptor [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2024.1434118/full]
- 22. Cancer biology and molecular genetics of A3 adenosine receptor [https://www.nature.com/articles/s41388-021-02090-z?error=cookies_not_supported&code=961e057f-dfab-4e2d-b168-b8bfd380cc48]
- 23. Adenosine A1 Receptor - an overview | ScienceDirect Topics [https://www.sciencedirect.com/topics/neuroscience/adenosine-a1-receptor]
- 24. Adenosine A1 and A2 Receptors: Structure-Function Relationships - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3448285/]
- 25. Adenosine A1 receptor - Wikipedia [https://en.wikipedia.org/wiki/Adenosine_A1_receptor]
- 26. , a target and regulator of estrogen receptorα... Adenosine A 1 receptor [https://www.nature.com/articles/onc2009409?error=cookies_not_supported&code=71dfb6e1-05bb-4322-b65c-585f50b5dbbe]
- 27. A2A/A2B Dual Antagonist M1069 Counteracts... Adenosine Receptor [https://pubmed.ncbi.nlm.nih.gov/39162025/]
- 28. Thieme E-Journals - Drug Research / Abstract [https://www.thieme-connect.de/products/ejournals/abstract/10.1055/a-2376-5771]
- 29. Adenosine receptors in breast cancer | Molecular Biology Reports [https://link.springer.com/article/10.1007/s11033-024-09382-z]
- 30. , Apoptosis , and proliferation in oral tissues. angiogenesis [https://pubmed.ncbi.nlm.nih.gov/10918211/]
- 31. Positive allosteric mechanisms of adenosine A1 receptor-mediated analgesia | Nature [https://www.nature.com/articles/s41586-021-03897-2]
- 32. of the Structure -bound human adenosine -Gi... adenosine A 1 receptor [https://pubmed.ncbi.nlm.nih.gov/29925945/]
- 33. : A review of risk factors and diagnosis - PMC Breast cancer [https://pmc.ncbi.nlm.nih.gov/articles/PMC10798762/]
- 34. treatment landscape for patients with locally recurrent... Current [https://breast-cancer-research.biomedcentral.com/articles/10.1186/s13058-019-1210-4]
- 35. RCSB PDB - 7LD3: Cryo-EM structure of the human adenosine ... [https://www.rcsb.org/structure/7LD3]
- 36. GEDI: An R Package for Integration of Transcriptomic Data from Multiple Platforms for Bioinformatics Applications - PubMed [https://pubmed.ncbi.nlm.nih.gov/39453042/]
- 37. Integration of quantitated expression estimates from polyA-selected and rRNA-depleted RNA-seq libraries - PubMed [https://pubmed.ncbi.nlm.nih.gov/28610557/]
- 38. Advances in the Applications of Bioinformatics and Chemoinformatics - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC10384252/]
- 39. Dynamic allosteric networks drive adenosine A1 receptor activation and G-protein coupling | eLife [https://elifesciences.org/articles/90773]
- 40. Adenosine A1 Receptor and Ligand Molecular Modeling - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10794910/]
- 41. , a target and regulator of ERα action... Adenosine A 1 receptor [https://pmc.ncbi.nlm.nih.gov/articles/PMC2829108/]
- 42. A Combination of Receptor-Based Pharmacophore Modeling & QM Techniques for Identification of Human Chymase Inhibitors | PLOS One [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0063030]
- 43. Structure-based pharmacophore modeling 1. Automated random pharmacophore model generation - ScienceDirect [https://www.sciencedirect.com/science/article/abs/pii/S109332632300027X]
- 44. Targeting human progesterone receptor (PR), through... [https://www.nature.com/articles/s41598-024-55321-0?error=cookies_not_supported&code=59a501d1-7f1b-4058-acc2-8a795bcc3c80]
- 45. Identification of phytomolecules as isoform and mutation specific... [https://bmcchem.biomedcentral.com/articles/10.1186/s13065-024-01317-w]
- 46. Drug design - Wikipedia [https://en.wikipedia.org/wiki/Drug_design]
- 47. Insights into the human A1 adenosine receptor from molecular dynamics simulation: structural study in the presence of lipid membrane | Medicinal Chemistry Research [https://link.springer.com/article/10.1007/s00044-015-1409-6]
- 48. Adenosine-A2A Receptor Pathway in Cancer Immunotherapy - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC8977492/]
- 49. The Story of MCF - 7 Breast Cancer Cell Line: 40 years of Experience in... [https://ar.iiarjournals.org/content/35/6/3147]
- 50. Overcoming high level adenosine-mediated immunosuppression by DZD2269, a potent and selective A2aR antagonist | Journal of Experimental & Clinical Cancer Research | Full Text [https://jeccr.biomedcentral.com/articles/10.1186/s13046-022-02511-1]
- 51. Estrogen alpha receptor antagonists for the treatment of breast ... [https://bmcchem.biomedcentral.com/articles/10.1186/s13065-018-0472-8]
- 52. : a web server for SwissTargetPrediction prediction of bioactive... target [https://pubmed.ncbi.nlm.nih.gov/24792161/]
- 53. SwissTargetPrediction: a web server for target prediction of bioactive small molecules - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4086140/]
- 54. Estrogen stimulates adenosine receptor expression subtypes in human breast cancer MCF-7 cell line - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC5772082/]
- 55. Adenosine and its receptors as therapeutic targets: An overview - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3744929/]
- 56. Structure of the Adenosine Reveals the Basis for... A 1 Receptor [https://pubmed.ncbi.nlm.nih.gov/28235198/]
- 57. (PDF) Structure of the Adenosine Reveals the Basis for... A 1 Receptor [https://www.academia.edu/110875800/Structure_of_the_Adenosine_A1_Receptor_Reveals_the_Basis_for_Subtype_Selectivity]
- 58. (PDF) Pharmacological characterization of adenosine A 2B receptors [https://www.academia.edu/8997507/Pharmacological_characterization_of_adenosine_A_2B_receptors]
- 59. Insights into the Adenosine Mechanistic Receptor’s Positive... A 1 [https://pmc.ncbi.nlm.nih.gov/articles/PMC11726717/]
- 60. Frontiers | Update on the recent development of allosteric modulators for adenosine receptors and their therapeutic applications [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2022.1030895/full]
- 61. Structural Basis for Binding of Allosteric Drug Leads in the Adenosine A1 Receptor - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6237911/]
- 62. Positive allosteric of mechanisms -mediated... adenosine A 1 receptor [https://www.nature.com/articles/s41586-021-03897-2?error=cookies_not_supported&code=54a6998e-8d8e-4470-8074-f3250d6eeb98]
- 63. Small molecule allosteric modulation of the adenosine A1 receptor - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10331460/]
- 64. Separation of on-target efficacy from adverse effects through rational design of a bitopic adenosine receptor agonist - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3970544/]
- 65. Mechanistic Insights into the Adenosine A1 Receptor's Positive Allosteric Modulation for Non-Opioid Analgesics - PubMed [https://pubmed.ncbi.nlm.nih.gov/39768211/]
- 66. The importance of adenosine in tumor... A 1 receptors [https://www.jcgtm.org/index.php/home/article/view/463]
- 67. Discovery and Structure-Activity Relationship Studies of Novel Adenosine A1 Receptor-Selective Agonists - PubMed [https://pubmed.ncbi.nlm.nih.gov/36270633/]
- 68. Discovery and Structure–Activity Relationship Studies of Novel Adenosine A1 Receptor-Selective Agonists - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9661479/]
- 69. Modulation of adenosine receptor affinity and intrinsic efficacy in adenine nucleosides substituted at the 2-position - ScienceDirect [https://www.sciencedirect.com/science/article/abs/pii/S0968089604002160]
- 70. High Affinity Acylating Antagonists for the A1 Adenosine Receptor: Identification of Binding Subunit - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3557832/]
- 71. Adenosine Metabolism, Immunity and Joint Health - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC5899962/]
- 72. Adenosine - Wikipedia [https://en.wikipedia.org/wiki/Adenosine]
- 73. The progress and prospects of targeting the adenosine pathway in... [https://biomarkerres.biomedcentral.com/articles/10.1186/s40364-025-00784-0]
- 74. Adenosine metabolism – emerging concepts for cancer therapy - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7224341/]
- 75. Frontiers | Metabolic Aspects of Adenosine Functions in the Brain [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2021.672182/full]
- 76. Adenosine Receptors and Cancer - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3598010/]
- 77. Chemoresistance mechanisms to 5-Fluorouracil and reversal strategies in lung and breast cancer | Scientific Reports [https://www.nature.com/articles/s41598-025-90532-z]
- 78. Microcurrent stimulation induces cell death in p53-mutant and 5-FU-resistant breast cancer - PubMed [https://pubmed.ncbi.nlm.nih.gov/40570957/]
- 79. Androgen Receptor as a Potential Target for Treatment of Breast Cancer - PubMed [https://pubmed.ncbi.nlm.nih.gov/28474005/]
- 80. Adenosine receptors as therapeutic targets | Nature Reviews Drug Discovery [https://www.nature.com/articles/nrd1983]
- 81. Adenosine Receptor Antagonists to Combat Cancer and to Boost Anti-Cancer Chemotherapy and Immunotherapy - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC8616521/]
- 82. Chemotherapy-induced adenosine A2B receptor expression mediates epigenetic regulation of pluripotency factors and promotes breast cancer stemness [https://www.thno.org/v12p2598.htm]
- 83. Chemotherapy-induced adenosine A2B receptor expression mediates epigenetic regulation of pluripotency factors and promotes breast cancer stemness - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8965495/]
- 84. The A2B adenosine receptor in MDA-MB-231 breast cancer cells diminishes ERK1/2 phosphorylation by activation of MAPK-phosphatase-1 - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6114864/]
- 85. Exploring neurotransmitters and their receptors for breast ... cancer [https://pmc.ncbi.nlm.nih.gov/articles/PMC9925324/]
- 86. of imidazothioxanthones in murine and human... Antitumor activity [https://pubmed.ncbi.nlm.nih.gov/15161044/]
- 87. Frontiers | BYS10, a novel selective RET inhibitor, exhibits potent... [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2025.1670140/full]
- 88. The progress and prospects of targeting the adenosine in... pathway [https://pmc.ncbi.nlm.nih.gov/articles/PMC12090549/]
- 89. in Triple-Negative Epigenetic —The... Alterations Breast Cancer [https://pmc.ncbi.nlm.nih.gov/articles/PMC7916242/]
- 90. Adenosine arrests breast cancer cell motility by A3 receptor stimulation - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC5124008/]
- 91. Frontiers | Distinct Roles of Adenosine Deaminase Isoenzymes... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2022.903461/full]