Molecular Alignment in 3D-QSAR: A Comparative Guide to Methods, Applications, and Best Practices for Drug Discovery
This article provides a comprehensive analysis of molecular alignment methodologies central to 3D-QSAR, a critical technique in modern computer-aided drug design.
Molecular Alignment in 3D-QSAR: A Comparative Guide to Methods, Applications, and Best Practices for Drug Discovery
Abstract
This article provides a comprehensive analysis of molecular alignment methodologies central to 3D-QSAR, a critical technique in modern computer-aided drug design. Aimed at researchers and drug development professionals, it explores foundational principles, from the role of molecular interaction fields (MIFs) and the probe concept to the crucial impact of alignment on model predictability. The review systematically compares manual, automated, and alignment-independent techniques, offering practical insights for method selection, troubleshooting common pitfalls, and validating models through statistical and prospective applications. By synthesizing traditional approaches with emerging trends, including AI integration, this guide serves as a strategic resource for optimizing 3D-QSAR workflows to enhance the efficiency and success of lead optimization and scaffold hopping in drug discovery projects.
The Cornerstone of 3D-QSAR: Understanding Why Molecular Alignment Matters
In modern drug discovery, understanding the interaction between a receptor and its ligand is a fundamental step in the rational design of new therapeutic agents. This process is inherently three-dimensional, as the biological receptor does not perceive a ligand as a simple set of atoms and bonds, but rather as a specific three-dimensional shape that carries a complex distribution of molecular forces [1]. This article will explore the principle of three-dimensional perception within the context of 3D Quantitative Structure-Activity Relationship (3D-QSAR) studies, with a specific focus on comparing the manual and automated molecular alignment methods that are critical to this process.
The 3D Basis of Molecular Recognition
Molecular binding is a three-dimensional event. The affinity of a ligand for its receptor is determined by the interplay of intermolecular forces—such as steric bulk, electrostatic potential, and hydrogen bonding—that depend entirely on the relative spatial orientation of the two molecules [1].
The receptor perceives a ligand through these interaction forces. At long distances, the electrostatic field, which can be calculated using Coulomb's law, guides the initial approach of the ligand. At shorter ranges, steric forces, often described by a Lennard-Jones potential, become dominant, controlling the final binding step by determining which shapes can fit without clash and where bulky groups might be accommodated [1]. This is why 3D-QSAR methods move beyond simple molecular descriptors (like logP) and instead represent molecules by calculating the values of these steric and electrostatic fields at numerous points in the space surrounding them [2] [1].
Visualizing Molecular Interaction Fields
To operationalize this principle, 3D-QSAR methods use the concept of Molecular Interaction Fields (MIFs). A MIF is measured by placing a conceptual "probe" atom (e.g., an sp3 carbon with a +1 charge for electrostatic fields) at various points on a 3D lattice or grid surrounding the molecule. The interaction energy between the molecule and the probe is calculated at each grid point, mapping out the regions of favorable and unfavorable interactions [1]. These fields can be visualized as iso-potential surfaces, providing researchers with an intuitive, three-dimensional map of the molecular forces that a receptor would "feel" [1].
The following diagram illustrates the core workflow for generating these critical molecular interaction fields.
Comparing Molecular Alignment Methodologies in 3D-QSAR
A critical step in 3D-QSAR is molecular alignment—the superimposition of all molecules in a dataset within a shared 3D reference frame that reflects their putative bioactive conformations [2]. The quality of this alignment directly impacts the model's predictive power. The two primary approaches, manual and automated alignment, were directly compared in a seminal study using 113 flexible cyclic urea inhibitors of HIV-1 protease [3].
Manual vs. Automated Alignment: An Experimental Comparison
The following table summarizes the key findings from this comparative study, which utilized Comparative Molecular Field Analysis (CoMFA) and Comparative Molecular Similarity Indices Analysis (CoMSIA) models.
Table 1: Quantitative Comparison of Manual vs. Automated Alignment in 3D-QSAR (HIV-1 Protease Inhibitor Study) [3]
| Metric | Manual Alignment | Automated Alignment | Experimental Context |
|---|---|---|---|
| Best Cross-Validated R² (q²) | Statistically higher values | 0.649 | Leave-One-Out (LOO) cross-validation on training set |
| Best Predictive R² | 0.754 | 0.754 | Predictive power on an external test set of inhibitors |
| Model Robustness | Lower | More robust | Ability to generalize predictions to new, unseen compounds |
| Alignment Basis | Known X-ray structures | Molecular docking into the target protein (HIV-1 PR) | Docked poses agreed with X-ray structural information |
| Key Identified Interactions | Hydrogen bonds with Gly48, Gly48', Asp30 backbone | Hydrogen bonds with Gly48, Gly48', Asp30 backbone | Both methods identified the same critical receptor-ligand interactions |
Key Insight: While manual alignment can yield models with slightly higher internal statistical scores, automated alignment based on molecular docking can produce more robust models for predicting the activities of an external inhibitor set [3]. This is a significant advantage in real-world drug discovery, where the goal is to predict the activity of novel compounds.
Detailed Experimental Protocols
To ensure reproducibility, this section outlines the core methodologies for the alignment and modeling techniques discussed.
Protocol 1: Manual Alignment for 3D-QSAR
The traditional manual approach relies heavily on researcher intuition and known structural data [2].
- Template Selection: Choose a reference molecule, often a high-affinity ligand with a experimentally determined (e.g., X-ray) 3D structure in its bioactive conformation.
- Common Substructure Identification: Identify the largest common substructure (e.g., a scaffold or pharmacophore) shared across the data set [2].
- Superimposition: Manually align all other molecules in the dataset onto the reference template by fitting the atoms of the common substructure. This is often done using visualization software.
- Field Calculation: With all molecules in a common orientation, proceed to calculate steric and electrostatic fields on a surrounding grid.
Protocol 2: Automated Docking-Based Alignment
This structure-based method leverages computational docking to define alignment [3] [4].
- Protein Preparation: Obtain the 3D structure of the target receptor (e.g., from Protein Data Bank). Prepare the structure by adding hydrogen atoms, assigning partial charges, and defining the binding site.
- Ligand Preparation: Generate low-energy 3D conformations for all ligands in the dataset and assign correct ionization states at physiological pH [5].
- Molecular Docking: Use a docking program (e.g., Glide) to predict the binding pose of each ligand within the defined binding site of the receptor. A grid box (e.g., 20 Å x 20 Å x 20 Å) is typically centered on the binding site [5].
- Pose Extraction and Alignment: Extract the highest-ranked docking pose for each ligand. These poses, all situated within the same receptor frame, automatically constitute the aligned dataset for subsequent 3D-QSAR analysis [3] [4].
The Scientist's Toolkit: Essential Reagents & Software for 3D-QSAR
Table 2: Key Research Tools and Resources for 3D-QSAR Studies
| Tool/Resource | Type | Primary Function in 3D-QSAR | Example Use Case |
|---|---|---|---|
| Sybyl (Tripos) | Software Suite | Molecular modeling, geometry optimization, and running classic 3D-QSAR methods like CoMFA and CoMSIA [2]. | Generating steric and electrostatic field descriptors from an aligned molecule set. |
| Schrödinger Suite (Glide) | Software Suite | Protein and ligand preparation, and high-performance molecular docking for automated alignment [5]. | Predicting the binding pose of a novel ligand in a receptor with a known crystal structure (e.g., Sigma1 receptor [5]). |
| GRID | Software | A structure-based program for calculating interaction fields using a wide variety of chemical probes [1]. | Identifying "hot spots" in a protein binding site that favor interactions with specific chemical groups (e.g., carbonyl oxygen, amine). |
| RDKit | Open-Software | Cheminformatics toolkit for converting 2D structures to 3D, conformer generation, and identifying maximum common substructures (MCS) [2]. | Automating the initial generation of 3D conformers for a large dataset of compounds. |
| QSAR Toolbox | Free Software | Profiling chemicals, finding analogues, and filling data gaps via read-across and (Q)SAR models [6]. | Screening a new chemical for potential endocrine disruption by profiling it against a database of known thyroid peroxidase (TPO) inhibitors [6]. |
The fundamental principle that receptors perceive ligands in three dimensions through their shape and interaction fields is the cornerstone of 3D-QSAR. The choice of molecular alignment method is critical for building predictive models. Evidence shows that while manual alignment can provide models with high internal consistency, automated docking-based alignment produces more robust models for predicting the activity of external compounds [3]. This makes automated methods highly valuable, particularly when a well-characterized protein structure is available. The integration of these alignment strategies with powerful computational tools allows researchers to translate the abstract concept of 3D perception into concrete, predictive models that accelerate rational drug design.
In the field of three-dimensional quantitative structure-activity relationship (3D-QSAR) research, Molecular Interaction Fields (MIFs) and the probe concept together form the foundational framework for comparing molecular properties and predicting biological activity. MIFs are three-dimensional interaction maps that describe the intermolecular interactions expected to form around target molecules [7]. The core principle underpinning MIF generation is the probe concept—the use of specific chemical groups to quantitatively measure the interaction potential around molecules of interest [1].
The biological activity of a ligand depends substantially on its affinity for its receptor, a process that occurs in three-dimensional space [1]. Since receptors perceive ligands not as collections of atoms but as shapes carrying complex interaction forces, MIFs provide a crucial computational approach to quantify these forces when the receptor structure is unknown [1]. This review provides a comprehensive comparison of MIF methodologies, probe types, and their applications in modern drug discovery, with particular emphasis on their role in comparing molecular alignment methods within 3D-QSAR research.
Theoretical Foundations: How MIFs and Probes Work
Basic Principles of Molecular Interaction Fields
Molecular Interaction Fields represent a computational method based on the analysis and comparison of three-dimensional molecular fields (steric, electrostatic, etc.) generated in the space surrounding chemical compounds [1]. The primary objective is to establish a statistical correlation between these fields and biological activities [1]. Unlike classical 2D-QSAR, which describes molecular properties using parameters independent of spatial coordinates (e.g., logP, molar refractivity), 3D-QSAR represents properties as sets of values of (x,y,z) functions measured at numerous locations in the surrounding space [1]. This fundamental difference results in significantly more molecular descriptors being available in 3D-QSAR compared to classical approaches.
The generation of MIFs relies on the systematic calculation of interaction energies between a target molecule and a probe positioned at numerous grid points within a three-dimensional lattice surrounding the molecule [1] [7]. This lattice-based sampling enables the computationally efficient characterization of spatial interaction patterns. The resulting interaction energy values at these grid points serve as descriptors for constructing quantitative models or visual contour maps that highlight regions of favorable or unfavorable interactions [7].
The Probe Concept: Measuring Molecular Fields
The probe concept is central to MIF generation, founded on the principle that a molecular interaction field can only be measured using an appropriate "receiver" capable of interacting with it [1]. Similar to how a compass detects Earth's magnetic field, molecular interaction fields require specialized probes for their detection and quantification.
Probes are chemical entities—ranging from single atoms to functional groups or entire molecules—that are systematically positioned at grid points surrounding the target structure [1]. At each point, the interaction energy between the target and the probe is calculated using potential energy functions, creating a comprehensive map of interaction potentials [1]. The selection of appropriate probes is crucial, as they must match the field type being measured (e.g., van der Waals probes for steric fields, charged probes for electrostatic fields) [1].
Table 1: Fundamental Probe Types and Their Applications in MIF Generation
| Probe Type | Chemical Representation | Primary Field Measured | Common Applications |
|---|---|---|---|
| Single Atom | Carbon sp³ | Steric field | Mapping molecular shape and steric hindrance [1] |
| Charged Atom | Carbon sp³ with +1 charge | Electrostatic field | Mapping electrostatic potential and charge distribution [1] |
| Functional Groups | CH₃, NH₂, CONH₂, OH | Specific functional interactions | Hydrogen bonding, hydrophobic interactions [1] |
| Whole Molecules | H₂O, NH₃⁺, COO⁻ | Complex interaction patterns | Solvation effects, ionic interactions [1] |
| Halogenated Probes | Chlorobenzene, Bromobenzene, Iodobenzene | Halogen bonding potential | σ-hole interactions in drug design [7] |
Key Methodologies and Probe Systems
Established MIF Generation Approaches
Several computational methodologies have been developed for generating and analyzing MIFs, each with distinctive probe systems and applications:
GRID Method: Developed by Peter Goodford in 1985, GRID was the first program based on MIF calculations [1]. This structure-based approach systematically explores binding sites by calculating interaction energies between a protein and various probes at each grid point [1]. The GRID force field employs a 6-4 potential function, which provides smoother energy calculations compared to the Lennard-Jones 6-12 potential used in early CoMFA methods [8]. The methodology offers dozens of specialized probes including single atoms, water, methyl groups, amine nitrogen, carbonyl oxygen, carboxylate, hydroxyl, and various metal cations (Na⁺, K⁺, Ca⁺⁺, Fe⁺⁺, Fe⁺⁺⁺, Zn⁺⁺, Mg⁺⁺) [1].
Comparative Molecular Field Analysis (CoMFA): As the first validated 3D-QSAR approach, CoMFA correlates biological activity with interaction energy contributions at every grid point surrounding a set of aligned molecules [8]. The method typically employs steric (Lennard-Jones potential) and electrostatic (Coulombic potential) probes [1]. CoMFA has become a prototype for 3D-QSAR methods and remains widely used despite the development of more advanced techniques [8].
Comparative Molecular Similarity Indices Analysis (CoMSIA): An extension of CoMFA, CoMSIA calculates molecular similarity indices from similarity fields and uses them as descriptors encoding steric, electrostatic, hydrophobic, and hydrogen-bonding properties [8]. This approach addresses some limitations of CoMFA by using a Gaussian function that avoids singularities and provides better differentiation of steric and electrostatic contributions.
Specialized Probe Developments
Recent methodological advances have focused on developing specialized probes for specific interaction types:
Halogen Bonding Probes: Conventional molecular mechanics models often fail to properly characterize halogen bonds due to their directional nature and the presence of σ-holes on halogen atoms [7]. Quantum mechanical (QM) calculations have emerged as the most reliable method for describing these interactions [7]. Recent research has employed chlorobenzene, bromobenzene, and iodobenzene as probes to map halogen-bond-formable areas around target molecules [7]. These QM-derived probes accurately reproduce the anisotropic nature of halogen interactions, which is crucial for modern structure-based drug design where halogenated compounds are increasingly common [7].
Knowledge-Based Approaches (SuperStar): The SuperStar method employs an alternative, empirical approach to generating 3D interaction maps using IsoStar—a knowledge-based library of intermolecular interactions constructed from the Cambridge Structural Database and Protein Data Bank [7]. Rather than calculating interaction energies, SuperStar predicts statistical probabilities of interactions around target molecules based on experimental data from crystal structures [7]. This approach provides complementary information to energy-based MIF calculations.
Table 2: Comparison of Major MIF Methodologies and Their Probe Systems
| Methodology | Probe Systems | Energy Functions | Key Advantages | Limitations |
|---|---|---|---|---|
| GRID [1] [8] | Extensive library: single atoms, functional groups, metal cations, molecular fragments | 6-4 potential function | Smooth energy calculations; diverse probe library; well-validated for active site analysis | Computational intensity for large systems |
| CoMFA [1] [8] | Standard steric and electrostatic probes (e.g., sp³ carbon, charged atoms) | Lennard-Jones (6-12) and Coulomb potentials | Established methodology; intuitive interpretation; high predictive ability | Sensitivity to molecular alignment; singularities near van der Waals surfaces |
| CoMSIA [8] | Similar to CoMFA with additional hydrophobic and H-bond probes | Gaussian-type distance-dependent functions | No singularities; better steric/electrostatic differentiation; additional field types | More parameters to optimize; potentially overfitted models |
| QM-MIF [7] | Halogenated benzene derivatives (Cl, Br, I) | Quantum mechanical (ωB97X-D, MP2) with BSSE correction | Accurate description of anisotropic interactions; reliable for halogen bonding | Extremely computationally intensive; limited to small systems without approximations |
| SuperStar [7] | Statistical distributions from database | Knowledge-based potentials from crystallographic data | Experimental basis; no force field parameterization required | Limited to well-represented interactions in databases |
Experimental Protocols and Workflows
Standard MIF Generation Protocol
The generation of Molecular Interaction Fields follows a systematic computational workflow:
Molecular Preparation: Target molecules are prepared with proper geometry optimization, protonation states, and conformation selection. For 3D-QSAR studies, molecules are typically aligned based on a common scaffold or pharmacophoric features [8].
Grid Definition: A three-dimensional lattice is superimposed around the target molecule(s), defining regularly spaced grid points where interaction energies will be calculated [1]. The grid dimensions and spacing are optimized to balance computational efficiency with adequate spatial resolution (typically 1-2 Å between grid points) [1].
Probe Selection: Appropriate probes are selected based on the chemical interactions of interest. Standard probes include steric (van der Waals), electrostatic, hydrophobic, and hydrogen bond donors/acceptors [1].
Energy Calculation: At each grid point, the interaction energy between the target and the probe is calculated using appropriate potential functions:
Data Analysis: The resulting energy matrices are analyzed using statistical methods, primarily Partial Least Squares (PLS) regression, to correlate field values with biological activities [8].
Advanced Protocol for Halogen Bond MIFs
Recent research has developed specialized protocols for mapping halogen bonding interactions using QM-based approaches [7]:
Diagram 1: Workflow for QM-Based Halogen Bond MIF Generation
Spherical Grid Setup: Define spherical grid points around the target molecule (e.g., N-methylacetamide as a protein main chain model) with radial points from 2-7 Å at 0.5 Å intervals, and polar/azimuth angles from 0°-180°/0°-360° at 10° intervals [7].
Probe Positioning: Place halogenated benzene probes (chlorobenzene, bromobenzene, iodobenzene) at each grid point with the halogen atom positioned directly on the point and the C-X bond axis aligned toward the target carbonyl oxygen [7].
QM Energy Calculation: Perform quantum mechanical calculations at the ωB97X-D/aug-cc-pVDZ-PP level for bromobenzene/iodobenzene systems or MP2/aug-cc-pVDZ for chlorobenzene systems, including counterpoise correction for basis set superposition error [7].
Energy Normalization: Normalize interaction energies to values between 0-1 based on the most stable energy encountered, setting repulsive interactions to 0 [7].
Function Approximation: Derive approximation functions Eₓ(r) for the MIFs using linear combinations of Gaussian functions to enable practical application to protein systems [7].
The Scientist's Toolkit: Essential Research Reagents and Probes
Table 3: Essential Research Reagents and Computational Probes for MIF Studies
| Reagent/Probe Type | Specific Examples | Primary Function | Application Context |
|---|---|---|---|
| Standard Steric Probes | sp³ Carbon atom | Maps steric hindrance and molecular shape | CoMFA, GRID studies of congeneric series [1] |
| Electrostatic Probes | +1 Charged carbon sp³ | Maps electrostatic potential around molecules | Identifying charge-assisted binding interactions [1] |
| Hydrogen Bond Probes | Carbonyl oxygen, amine nitrogen, hydroxyl group | Characterizes H-bond donor/acceptor properties | Predicting specific protein-ligand interactions [1] |
| Halogen Bond Probes | Chlorobenzene, Bromobenzene, Iodobenzene | Maps σ-hole interactions and directional preferences | Design of halogenated drugs with improved affinity [7] |
| Solvation Probes | Water molecule | Models hydrophobic effects and solvation/desolvation | Predicting binding thermodynamics and solubility [1] |
| Hydrophobicity Probes | DRY probe (in GRID) | Characterizes hydrophobic interaction regions | ADMET profiling and membrane permeability prediction [8] |
| Metal Coordination Probes | Na⁺, K⁺, Ca⁺⁺, Zn⁺⁺ cations | Maps metal-binding regions in proteins | Metalloenzyme inhibitor design and toxicology assessment [1] |
| Knowledge-Based Probes | Statistical distributions from crystallographic databases | Empirical interaction potentials from structural data | Complementary validation of force-field based MIFs [7] |
Comparative Analysis of Probe Performance
Field Type Comparisons and Applications
Different probe types generate distinct field information that illuminates various aspects of molecular recognition:
Steric Fields: Generated using van der Waals probes (typically carbon sp³), steric fields map shape complementarity and steric hindrance effects [1]. The repulsive component dominates at short distances due to electronic cloud interpenetration, while weak attractive dispersion forces operate at longer ranges [1]. These fields are particularly important for understanding selectivity issues in drug design.
Electrostatic Fields: Calculated using Coulomb's law with charged probes, electrostatic fields capture long-range charge-charge and dipole-dipole interactions that often guide initial ligand approach to binding sites [1]. Since the electrostatic potential decays with 1/r distance dependence (compared to 1/r¹² for steric repulsion), electrostatic effects operate over much longer distances than steric effects [1].
Hydrogen Bonding Fields: Specialized probes containing hydrogen bond donors (e.g., amine nitrogen) or acceptors (e.g., carbonyl oxygen) map the directionality and strength of hydrogen bonding interactions [1]. These fields are crucial for understanding specific molecular recognition in biological systems.
Halogen Bonding Fields: Using halogenated benzene probes, these fields capture the anisotropic nature of halogen atoms, particularly the σ-hole region along the C-X bond axis where favorable interactions with electron donors occur [7]. The strength of these interactions follows the trend I > Br > Cl > F, correlating with σ-hole size and polarizability [7].
Methodological Performance in Predictive Modeling
The effectiveness of different MIF approaches varies significantly across application domains:
Traditional CoMFA vs. Modern Methods: While CoMFA remains widely used, newer approaches like L3D-PLS (CNN-based Partial Least Squares) have demonstrated superior performance in certain applications. In 30 publicly available pre-aligned molecular datasets, L3D-PLS outperformed traditional CoMFA, highlighting the potential of machine learning approaches to extract more meaningful features from molecular interaction fields [9].
Computational Efficiency Considerations: Standard molecular mechanics-based MIF calculations offer practical computation times suitable for high-throughput screening, while QM-based approaches provide higher accuracy at substantially increased computational cost [7]. The development of approximation functions for QM-level MIFs represents a promising approach to balancing accuracy and efficiency [7].
Alignment Sensitivity: A significant limitation of many MIF approaches is their sensitivity to molecular alignment, with small alignment variations potentially causing substantial changes in field patterns and resulting QSAR models [8]. Methods like GRIND (Grid-Independent Descriptors) attempt to address this by using alignment-independent descriptors derived from MIFs [8].
Molecular Interaction Fields and the probe concept continue to evolve as essential tools in computational drug discovery. The integration of MIF methodologies with other computational approaches—particularly molecular docking and machine learning—represents a powerful trend in modern drug design [8]. As the field advances, we observe several promising developments: the creation of more specialized probes for under-represented interaction types, improved QM/MM hybrid approaches for accurate yet efficient field calculations, and the incorporation of deep learning architectures to extract complex patterns from high-dimensional MIF data [7] [9].
The continued refinement of MIF methodologies and probe systems will enhance our ability to compare molecular properties, predict biological activities, and ultimately accelerate the discovery of novel therapeutic agents. For researchers engaged in 3D-QSAR studies, thoughtful selection of appropriate probes and MIF generation methods remains crucial for obtaining meaningful, predictive models that effectively guide chemical optimization efforts.
The Critical Impact of Alignment on Predictive Model Accuracy and Interpretability
In the field of 3D Quantitative Structure-Activity Relationship (3D-QSAR) modeling, molecular alignment is a foundational step that critically influences the predictive accuracy and interpretability of computational models. This guide objectively compares different molecular alignment methods, supported by experimental data, to inform researchers and drug development professionals in their methodological selections.
Quantitative Comparison of Alignment Method Performance
Experimental data from diverse studies demonstrate that the choice of alignment strategy directly impacts key model performance metrics, including predictive correlation (R²) and computational efficiency.
Table 1: Performance Comparison of Different Molecular Alignment Strategies in 3D-QSAR
| Alignment Method | Dataset / Context | Key Performance Metrics | Reported Advantages | Reported Limitations |
|---|---|---|---|---|
| 2D-to-3D Direct Conversion (No Alignment) | 146 Androgen Receptor Binders [10] | R²Test = 0.61; Achieved in 3-7% of the time required by other methods [10]. | High speed; Avoids alignment subjectivity; Suitable for fairly inflexible substrates [10]. | May not be suitable for highly flexible molecules; Conformations not systematically reproducible [10]. |
| Bioactive Conformation (from PDB) | 461 Structures across 6 protein-ligand series [11] | Models combining 2D + 3D descriptors performed best, coding complementary molecular properties [11]. | Represents physiologically relevant binding geometry; High information content for descriptors [11]. | Dependent on availability of high-quality crystal structures; Does not account for protein flexibility. |
| Energy-Minimized Global Minimum | 146 Androgen Receptor Binders [10] | R²Test = 0.56 to 0.61 (range) [10]. | Provides consistent, reproducible geometries based on molecular thermodynamics [10]. | Computationally intensive; The global minimum may not represent the bioactive conformation [10]. |
| Template-Based Alignment | 146 Androgen Receptor Binders [10] | Performance was inferior to the 2D>3D model for the dataset studied [10]. | Can enforce a presumed biologically relevant orientation based on a known active molecule [2]. | Highly sensitive to the choice of template; Incorrect template leads to model failure [2]. |
| Consensus Predictions (Aggregate Models) | 146 Androgen Receptor Binders [10] | Consensus R²Test = 0.65 (superior to any single conformation method) [10]. | Mitigates risk of poor performance from any single, incorrect conformation strategy [10]. | Highest computational cost; Requires building and validating multiple models [10]. |
Detailed Experimental Protocols for Alignment Methods
The quantitative data in Table 1 were generated through rigorous experimental designs. Below are detailed methodologies for key alignment approaches cited in this guide.
Protocol 1: 2D-to-3D Direct Conversion and 3D-QSDAR Modeling
This protocol outlines the alignment-independent technique used in the androgen receptor binder study [10].
- Data Curation: A diverse dataset of 146 compounds with known binding affinity (log(RBA)) for the androgen receptor was assembled. Structural diversity was ensured, encompassing steroids, pesticides, phenols, and other classes [10].
- Conformation Generation: 2D structures were directly converted to 3D coordinates using molecular mechanics as implemented in Jmol, without further energy optimization or alignment. These are referred to as "2D > 3D" structures [10].
- Descriptor Calculation (3D-QSDAR Fingerprint): A unique "fingerprint" was constructed for each molecule from the NMR chemical shifts of all carbon atom pairs (X- and Y-axes) and the inter-atomic distances between each pair (Z-axis). This fingerprint represents electronic and steric qualities [10].
- Grid Generation and Binning: The 3D-SDAR parametric space was tessellated into regular grids (binning). For a given molecule, descriptors were generated based on the number of fingerprint elements belonging to each bin [10].
- Model Building and Validation: An ensemble modeling PLS algorithm performing multiple training/hold-out test randomization cycles was used to build averaged "composite" models. Predictive performance was evaluated on external test sets, yielding the R²Test values [10].
Protocol 2: Comparative Molecular Field Analysis (CoMFA) with Rigorous Alignment
This protocol details the standard workflow for alignment-dependent 3D-QSAR methods, such as CoMFA [2].
- Data Collection and 3D Structure Generation: A homogenous dataset of compounds with experimentally determined biological activities (e.g., IC50) is assembled. 2D structures are converted to 3D and geometry-optimized using force fields (e.g., UFF) or quantum mechanical methods to achieve low-energy conformations [2].
- Molecular Alignment:
- Template Selection: A known active compound or a maximum common substructure (MCS) is chosen as a template, assuming a shared binding mode [2].
- Superposition: All molecules are systematically superimposed onto the template within a shared 3D coordinate system. This can be achieved using algorithms like RMSD fitting to the MCS [2].
- Descriptor Calculation (Field Analysis): A 3D grid is created to encompass all aligned molecules. A probe atom (e.g., sp³ carbon with +1 charge) is placed at each grid point, and steric (Lennard-Jones) and electrostatic (Coulombic) interaction energies with each molecule are calculated [2].
- Model Building and Visualization: Partial Least Squares (PLS) regression is used to correlate the field descriptors with biological activity. The model is visualized as 3D contour maps, showing regions where specific steric or electrostatic features enhance or diminish activity [2].
Protocol 3: Building a Model Using Bioactive Conformations from PDB
This protocol describes the curation of a dataset with experimentally determined bioactive conformations for a robust comparison of 2D vs. 3D descriptors [11].
- Dataset Curation: The Protein Data Bank (PDB) was mined for sets of protein-ligand complexes sharing the same protein, with uniform activity data reported. This resulted in a carefully curated dataset of 461 structures across six series [11].
- Ligand Conformation Extraction: The 3D structure of each ligand was extracted directly from its protein-ligand complex crystal structure. This conformation is defined as the "bioactive conformation" [11].
- Descriptor Calculation and Modeling: For each ligand in its bioactive conformation, multiple classes of descriptors were computed: 2D descriptors, 3D descriptors, and a combination of 2D+3D descriptors. Models were built using multiple machine learning algorithms (k-Nearest Neighbors, Random Forest, Lasso Regression) [11].
- Validation: Model performances were rigorously evaluated on external test sets, which were derived from the parent dataset either randomly or in a rational manner [11].
Workflow Visualization: 3D-QSAR Model Building and Alignment
The following diagram illustrates the critical decision points and pathways in a 3D-QSAR workflow, highlighting the role of molecular alignment.
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful 3D-QSAR studies rely on a suite of software tools and computational resources for structure handling, alignment, and model building.
Table 2: Essential Research Reagents and Software Solutions for 3D-QSAR
| Item / Software | Function in 3D-QSAR | Relevance to Alignment |
|---|---|---|
| Sybyl-X | Comprehensive molecular modeling suite. | Used for structure optimization, molecular alignment, and performing CoMFA/CoMSIA studies [12]. |
| RDKit | Open-source cheminformatics toolkit. | Used for 2D to 3D structure conversion, maximum common substructure (MCS) search, and scaffold-based alignment [2]. |
| Jmol | Open-source Java viewer for 3D chemical structures. | Can be used for basic 2D to 3D molecular structure conversion without energy minimization [10]. |
| Protein Data Bank (PDB) | Database of experimentally determined 3D structures of proteins and nucleic acids. | Source of bioactive conformations of ligands for alignment or model validation [11]. |
| Select KBest | Feature selection algorithm. | Used to identify the most relevant 2D or 3D descriptors from a large pool before model building, improving model robustness [13]. |
| PLS Regression | Statistical method (Partial Least Squares). | Standard technique for building the QSAR model, capable of handling the high number of correlated 3D descriptors generated [2] [10]. |
In three-dimensional quantitative structure-activity relationship (3D-QSAR) modeling, molecular alignment constitutes the foundational step that significantly determines the success and predictive power of the resulting models. Unlike traditional 2D-QSAR methods that utilize numerical descriptors derived from molecular graphs, 3D-QSAR techniques incorporate the spatial orientation and three-dimensional characteristics of molecules, making the alignment process—the superposition of molecules in a shared 3D coordinate system—a critical determinant of model quality [2] [14]. The central challenge lies in reproducing the putative bioactive conformation and orientation that molecules adopt when interacting with their biological target, a process that requires careful consideration of molecular flexibility, conformational space, and pharmacophoric features [15].
The sensitivity of 3D-QSAR to alignment quality stems from its direct impact on molecular field calculations. Techniques such as Comparative Molecular Field Analysis (CoMFA) and Comparative Molecular Similarity Indices Analysis (CoMSIA) generate descriptors by measuring interaction energies or similarity indices at grid points surrounding the aligned molecules [16] [2]. Incorrect alignments introduce noise into these descriptors, compromising the model's ability to capture genuine structure-activity relationships. As noted by experts, "The majority of the signal is in the alignments, so you need to get those right. If your alignments are incorrect your model will have limited or no predictive power" [14]. This review systematically categorizes and evaluates the predominant alignment methodologies employed in contemporary 3D-QSAR research, providing a structured framework for selecting appropriate strategies based on specific research contexts.
Manual and Knowledge-Driven Alignment Methods
Core Scaffold Alignment
The most traditional alignment approach relies on identifying and superimposing common structural frameworks present across molecules in a dataset. This method is particularly effective for congeneric series where compounds share a recognizable rigid core, such as the steroid nucleus used in the seminal CoMFA study [17] [15]. The process typically involves selecting a reference molecule—often the most active compound or one with confirmed bioactive conformation—and aligning all other molecules to it by fitting atoms of the common scaffold [2] [14].
Manual alignment can be enhanced through maximum common substructure (MCS) identification, which algorithmically determines the largest shared structural fragment across molecules, even when explicit scaffolds are not immediately apparent [2]. This approach accommodates greater chemical diversity while maintaining a rational basis for superposition. Tools like RDKit's AllChem.ConstrainedEmbed() can generate 3D conformations that match scaffold atoms to a reference, ensuring consistent orientation across molecules [2]. Although this method reduces subjectivity compared to purely visual alignment, it remains dependent on the assumption that the common substructure defines the primary binding orientation.
Structure-Based Alignment Using Crystallographic Data
When experimental structural information is available, alignment based on protein-ligand complexes provides a biologically relevant reference frame. This approach utilizes crystallographic data of ligand-receptor complexes to derive template conformations for alignment [18] [15]. For example, in a 3D-QSAR study on NAMPT inhibitors, researchers used molecular docking to generate alignments based on predicted binding modes, which "produce an appropriate inhibitor conformation and alignment that yields 3D-QSAR models of comparable statistical quality as manual alignment" [18].
The principal advantage of structure-based alignment lies in its biological plausibility, as it explicitly accounts for complementarity with the target binding site. However, this method requires either experimental complex structures or reliable homology models, which may not be available for all targets. Additionally, the approach assumes consistent binding modes across the entire compound series, which may not hold for structurally diverse ligands.
Field-Based and Similarity-Driven Alignment Approaches
Field-Based Similarity Searching (FBSS)
Field-based methods represent a significant advancement in alignment techniques by utilizing molecular property fields rather than atomic positions as the basis for superposition. The FBSS algorithm positions molecules to maximize the similarity of their steric, electrostatic, and hydrophobic fields [17] [19]. This approach recognizes that structurally diverse molecules may share similar interaction potential with biological targets despite different atomic connectivity.
The methodology involves positioning each molecule at the center of a 3D grid and calculating molecular field values at each grid point [17]. Similarity between molecules is then computed using metrics such as Carbo similarity indices or Hodgkin similarity indices [17]. Comparative studies demonstrate that "the QSAR models resulting from the FBSS alignments are broadly comparable in predictive performance with the models resulting from manual alignments" [17] [19], validating the utility of this automated approach.
Gaussian Field-Based Alignment
Modern implementations of field-based alignment often employ Gaussian functions to calculate molecular similarity, offering several advantages over traditional potential-based methods. Gaussian functions produce continuous molecular similarity maps that avoid the abrupt, non-physical cutoffs observed in some CoMFA models [16] [20]. This continuity makes the alignment process less sensitive to minor conformational variations and grid positioning [16].
In practice, Gaussian field-based alignment computes steric, electrostatic, hydrophobic, hydrogen bond donor, and hydrogen bond acceptor fields using Gaussian-type functions [20]. For example, Schrödinger's Field-based QSAR tool employs "Gaussian-based electrostatic, steric, hydrogen bond donor (HBD), hydrogen bond acceptor (HBA) and hydrophobic potential fields" for molecular alignment and subsequent QSAR model development [20]. The smooth nature of these fields enhances alignment stability, particularly for datasets with significant conformational flexibility.
Figure 1: Workflow for Field-Based Molecular Alignment. This process generates multiple conformers, calculates molecular fields, and optimizes their similarity to produce aligned molecules for 3D-QSAR analysis.
Automated and Algorithmic Alignment Strategies
Pharmacophore-Based Automated Alignment
Pharmacophore-based automation represents a sophisticated approach that eliminates manual intervention by identifying common three-dimensional pharmacophoric features across active molecules. Tools like AutoGPA automatically generate "pharmacophore queries" – common 3D arrangements of features such as hydrogen bond acceptors, donors, hydrophobic areas, and charged groups – that induce optimal overlay of bioactive molecules [15]. The software exhaustively searches for pharmacophore queries that distinguish actives from inactives and uses these for both conformation selection and molecular alignment [15].
The AutoGPA workflow involves multiple stages: generating low-energy conformations for each molecule, assigning pharmacophore features to each conformation, identifying common 3D pharmacophore arrangements, and selecting the alignment that produces the best 3D-QSAR model statistics [15]. Validation studies demonstrate that this automated approach can achieve predictive performance comparable to manual methods, with the significant advantage of objectivity and reproducibility. In one case study, AutoGPA generated models with q² = 0.76 and r² = 0.91, outperforming traditional CoMFA while requiring no prior knowledge of bioactive conformations [15].
Alignment-Independent 3D-QSAR Methods
For specific applications, alignment-independent techniques offer an alternative that bypasses the alignment challenge entirely. 3D-Spectral Data-Activity Relationship (3D-SDAR) represents one such method that uses NMR chemical shifts and interatomic distances to create unique molecular "fingerprints" without requiring molecular superposition [10]. This technique tessellates the 3D-SDAR space into regular grids, converting fingerprint information into descriptors that capture both electronic and steric properties while remaining inherently alignment-free [10].
Surprisingly, studies comparing 3D-SDAR models built from carefully energy-minimized conformations versus simple 2D-to-3D converted structures found that the latter "produced R²Test = 0.61" and "was superior to energy-minimized and conformation-aligned models and was achieved in only 3–7% of the time required using the other conformation strategies" [10]. This suggests that for certain nuclear receptor targets, where strong activities are produced by fairly inflexible substrates, simplified approaches can yield satisfactory results with dramatically reduced computational overhead.
Comparative Analysis of Alignment Methodologies
Performance Metrics Across Alignment Strategies
The effectiveness of alignment methods can be quantitatively assessed through statistical parameters of resulting 3D-QSAR models, including cross-validated correlation coefficient (q²), conventional correlation coefficient (r²), and predictive performance on external test sets. The table below summarizes comparative performance data for different alignment strategies applied to various biological systems.
Table 1: Comparative Performance of Different Alignment Methods in 3D-QSAR Studies
| Alignment Method | Biological System | q² | r² | Test Set Prediction r² | Reference |
|---|---|---|---|---|---|
| Field-Based Similarity Searching (FBSS) | Steroids (CBG) | 0.65 | 0.89 | Comparable to manual | [17] |
| Field-Based Similarity Searching (FBSS) | Acetylcholinesterase inhibitors | 0.55 | 0.94 | Comparable to manual | [17] |
| Pharmacophore-Based (AutoGPA) | PDK1 inhibitors | 0.76 | 0.91 | 0.65 | [15] |
| Docking-Based Alignment | NAMPT inhibitors | - | 0.84 | 0.85 | [18] |
| 2D-to-3D Conversion (3D-SDAR) | Androgen receptor binders | - | - | 0.61 | [10] |
Strategic Selection Guidelines
Choosing an appropriate alignment strategy requires careful consideration of multiple factors, including dataset characteristics, available structural information, and computational resources. The following guidelines emerge from comparative studies:
- For congeneric series with rigid cores: Manual scaffold-based alignment often suffices, particularly when supported by crystallographic data or reliable docking poses [2] [14].
- For structurally diverse datasets: Field-based or pharmacophore-based automated methods generally outperform manual approaches by identifying non-obvious yet biologically relevant superpositions [17] [15].
- When binding mode consistency is uncertain: Consensus approaches utilizing multiple alignment strategies or conformation-independent methods may provide more robust models [10] [14].
- For large-scale screening applications: Simplified 2D-to-3D conversion or alignment-free methods offer practical efficiency with acceptable predictive performance for specific target classes [10].
Notably, the pursuit of optimal alignment must be disciplined to avoid statistical overfitting. As cautioned by experienced practitioners, "you must not change the X data while paying attention (either directly or indirectly) to the Y data (the activities)" [14]. Alignment refinement based on model statistics constitutes circular reasoning and produces invalid models with artificially inflated performance metrics.
Essential Research Reagents and Computational Tools
Table 2: Key Software Tools for Molecular Alignment in 3D-QSAR Research
| Tool/Software | Alignment Approach | Key Features | Accessibility |
|---|---|---|---|
| Schrödinger Field-Based QSAR | Gaussian field-based | Five molecular field types (steric, electrostatic, HBD, HBA, hydrophobic); docking-based alignment | Commercial |
| Py-CoMSIA | User-defined | Open-source Python implementation; compatible with RDKit for conformer generation | Open-source [16] |
| AutoGPA | Pharmacophore-based | Automatic pharmacophore elucidation; conformation selection and alignment | Commercial [15] |
| FBSS | Field-based similarity | Field-based molecular similarity optimization; automated alignment | Research implementation [17] |
| Cresset Forge/Torch | Field-based and scaffold-based | Combined substructure and field similarity alignment; multiple reference molecules | Commercial [14] |
| 3D-QSDAR | Alignment-independent | Uses NMR chemical shifts and interatomic distances; no molecular superposition required | Research implementation [10] |
Molecular alignment remains both a challenge and opportunity in 3D-QSAR modeling. While manual methods continue to offer intuitive appeal for congeneric series, automated approaches based on field similarity and pharmacophore perception provide robust alternatives that reduce subjectivity and accommodate chemical diversity. Emerging open-source implementations such as Py-CoMSIA promise to increase accessibility to advanced 3D-QSAR methodologies [16], while alignment-independent techniques offer practical solutions for specific applications where traditional alignment proves problematic.
The critical consideration across all methodologies is maintaining alignment objectivity—the superposition must reflect plausible binding modes without being unduly influenced by activity data. As the field advances, integration of machine learning with physics-based alignment methods may further enhance predictive performance while reducing manual intervention. Regardless of methodological innovations, the foundational principle endures: in 3D-QSAR, alignment quality ultimately determines model success, making thoughtful selection and execution of alignment strategies essential for meaningful structure-activity insights.
A Practical Guide to 3D-QSAR Alignment Techniques and Their Real-World Applications
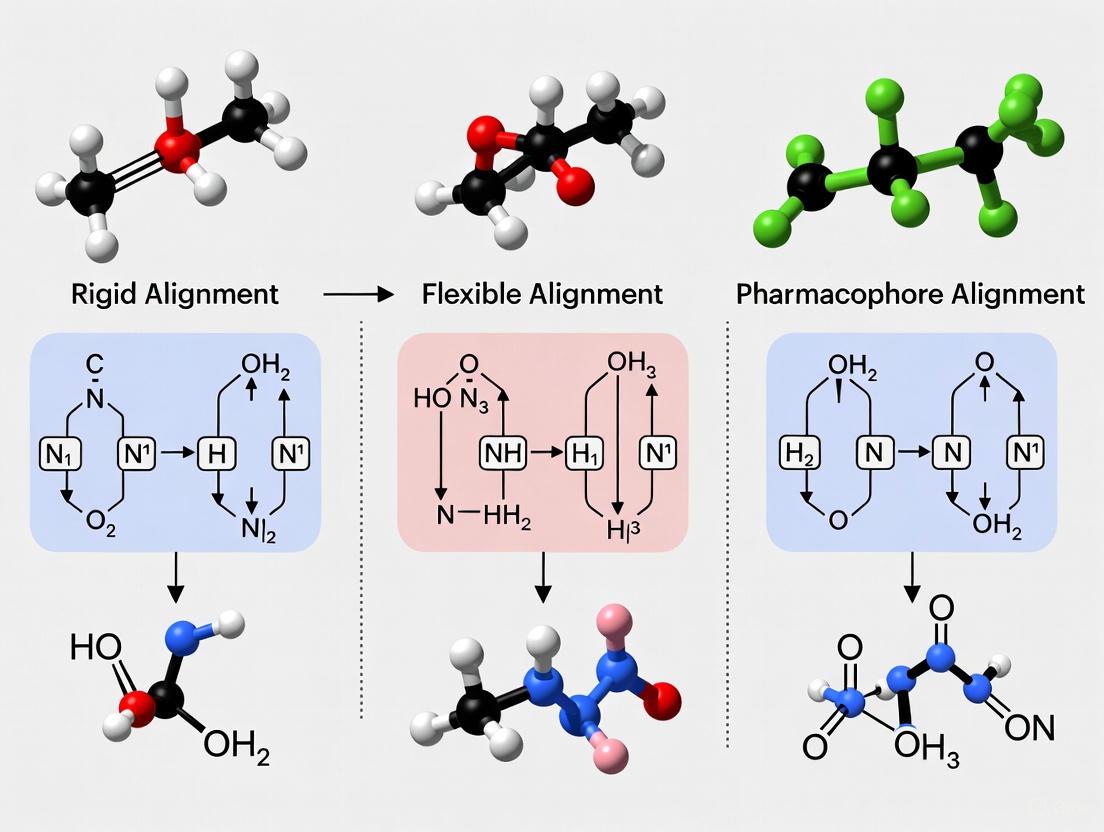
In the realm of three-dimensional quantitative structure-activity relationship (3D-QSAR) studies, the alignment of molecules is a critical step that predates the extraction of meaningful biological insights. Two predominant ligand-based methodologies—pharmacophore mapping and common scaffold alignment—serve as foundational approaches for superimposing molecules based on distinct principles. Pharmacophore mapping involves the spatial alignment of molecules based on their essential functional features—such as hydrogen bond donors, acceptors, and hydrophobic regions—rather than their atomic backbone. This method abstracts a molecule into a set of steric and electronic features necessary for its biological interaction [21] [22]. In contrast, common scaffold alignment relies on identifying and superimposing a shared, often rigid, structural framework or maximum common substructure (MCS) present across a set of active compounds [23]. The choice between these methodologies directly influences the predictive power and interpretability of subsequent 3D-QSAR models, guiding researchers in understanding the key structural determinants of biological activity for drug discovery.
Methodological Principles and Experimental Protocols
Core Concepts of Pharmacophore Mapping
A pharmacophore is defined by the International Union of Pure and Applied Chemistry (IUPAC) as "the ensemble of steric and electronic features that is necessary to ensure the optimal supramolecular interactions with a specific biological target structure and to trigger (or to block) its biological response" [21]. This approach distills molecular recognition into a three-dimensional arrangement of abstract features representing interaction types, moving beyond specific functional groups. The core features include:
- Hydrogen-bond donors (HBD) and acceptors (HBA): Represented as vectors indicating the direction of potential hydrogen bonds.
- Hydrophobic (H) and Aromatic (AR) features: Representing areas for van der Waals and dispersion interactions.
- Charged features: Positively (PO) or negatively (NE) charged centers for electrostatic interactions.
- Exclusion volumes (XVol): Representing regions where steric clashes would prevent binding, thereby mimicking the topology of the binding pocket [21] [22].
The experimental workflow for pharmacophore model generation follows two primary approaches: structure-based and ligand-based. Structure-based pharmacophore modeling utilizes experimentally determined protein-ligand complexes (e.g., from X-ray crystallography or NMR stored in the Protein Data Bank) to extract the interaction pattern directly from the binding site [21]. Software tools like Discovery Studio and LigandScout can generate pharmacophore features directly from the binding site topology, even in the absence of a bound ligand [21]. Ligand-based pharmacophore modeling, conversely, addresses the absence of a known receptor structure by identifying common feature patterns from a set of active, conformationally diverse ligands. This method requires the alignment of multiple active compounds to identify their shared pharmacophoric elements [22].
Core Concepts of Common Scaffold Alignment
Common scaffold alignment, often implemented through Maximum Common Substructure (MCS) algorithms, operates on the principle that structurally similar compounds, particularly those sharing a core framework, are likely to exhibit similar biological activities. This methodology involves:
- Identification of a shared structural core: The algorithm identifies the largest common chemical substructure present across all or most molecules in the dataset.
- Conformational sampling: Generating biologically relevant, low-energy 3D conformations for each molecule, typically within a specified energy window (e.g., 2.5 kcal/mol) [23].
- Structural superposition: Aligning molecules by fitting their identified common scaffold, often using rigid-body rotations and translations to minimize the root-mean-square deviation (RMSD) between matched atoms [23].
The MCS alignment is particularly valuable when working with congeneric series of compounds—molecules derived from a common chemical scaffold with variations at specific substituent positions. The quality of the alignment is highly sensitive to the accuracy of the conformational analysis and the correctness of the identified common substructure.
Workflow Visualization
The diagram below illustrates the comparative workflows for pharmacophore mapping and common scaffold alignment, highlighting their distinct logical pathways from input data to final 3D-QSAR model input.
Performance Comparison and Experimental Data
Virtual Screening Performance
Virtual screening represents a critical application where the performance of alignment methods can be quantitatively evaluated. The table below summarizes key performance metrics reported for pharmacophore-based and scaffold-based approaches.
Table 1: Virtual Screening Performance Comparison
| Method | Application Context | Reported Hit Rate | Enrichment Factor | Key Strengths |
|---|---|---|---|---|
| Pharmacophore Mapping | Various target-based screening campaigns [21] | 5-40% | Significantly higher than random screening | Identifies structurally diverse hits (scaffold hopping) |
| Common Scaffold/Similarity | Conventional similarity searching [24] | Typically <1% for random selection; varies with similarity threshold | Lower than pharmacophore methods | Effective for lead optimization in congeneric series |
Pharmacophore-based virtual screening consistently demonstrates superior hit rates compared to traditional methods. For instance, while random screening or simple similarity searching typically yields hit rates below 1% (e.g., 0.55% for glycogen synthase kinase-3β, 0.075% for PPARγ), pharmacophore-based approaches routinely achieve hit rates between 5-40% in prospective studies [21]. This significant enhancement stems from the method's ability to capture essential interaction patterns rather than structural similarity alone.
Predictive Accuracy in 3D-QSAR Modeling
The alignment method directly impacts the statistical quality and predictive power of resulting 3D-QSAR models. Recent studies comparing different alignment strategies in specific drug discovery contexts reveal distinct performance patterns.
Table 2: 3D-QSAR Model Performance with Different Alignment Rules
| Alignment Method | Target | Statistical Performance (q²/r²) | Key Advantages | Reference Case |
|---|---|---|---|---|
| Pharmacophore-Based | SARS-CoV-2 Mpro inhibitors [23] | q² = 0.81, r² = 0.71 (Field 3D-QSAR) | Identifies key interaction regions; explains activity cliffs | Field 3D-QSAR model [23] |
| Common Scaffold (MCS) | SARS-CoV-2 Mpro inhibitors [23] | High predictive accuracy (model dependent) | Works well with congeneric series; intuitive alignment | MCS-based alignment in Flare [23] |
| Knowledge-Guided Diffusion (DiffPhore) | Generalized pharmacophore mapping [25] [26] | State-of-the-art pose prediction | Handles flexibility; incorporates directional constraints | DiffPhore framework [25] |
In a direct comparison study on SARS-CoV-2 main protease (Mpro) inhibitors, both alignment methods produced robust 3D-QSAR models. The pharmacophore-based Field 3D-QSAR model demonstrated strong predictive power with a q² of 0.81 and r² of 0.71, comparable to the best common scaffold-based models [23]. However, the pharmacophore approach offered the distinct advantage of visualizing regions where electrostatic and steric effects strongly influenced activity, thereby providing clearer guidance for molecular optimization.
Scaffold Hopping Potential
Scaffold hopping—the identification of novel core structures with similar biological activity—represents a critical test for the ability of alignment methods to transcend structural similarity. Pharmacophore mapping excels in this domain because it focuses on functional requirements rather than structural frameworks. By abstracting molecules to their essential features, pharmacophore models can identify structurally distinct compounds that fulfill the same interaction pattern [24]. In contrast, common scaffold alignment, by its nature, prioritizes structural conservation and is less suited for scaffold hopping unless the common substructure is defined very loosely, potentially at the cost of alignment quality. Modern AI-driven molecular representation methods that build upon pharmacophore principles have further enhanced scaffold hopping capabilities by enabling more flexible exploration of chemical space [24].
The Scientist's Toolkit: Essential Research Reagents and Solutions
Successful implementation of pharmacophore mapping and scaffold alignment requires specialized software tools and computational resources. The table below catalogues key solutions utilized in the field.
Table 3: Essential Research Reagent Solutions for Molecular Alignment
| Tool/Solution | Type | Primary Function | Application Context |
|---|---|---|---|
| LigandScout [21] | Software | Structure-based & ligand-based pharmacophore modeling | Virtual screening, binding site analysis |
| Discovery Studio [21] | Software | Pharmacophore model generation from binding sites | CADD, structure-based design |
| Flare [23] | Software | MCS alignment and Field 3D-QSAR | Molecular docking, 3D-QSAR studies |
| AncPhore [25] [26] | Software | Pharmacophore tool for dataset generation | Creation of 3D ligand-pharmacophore pairs |
| DiffPhore [25] [26] | AI Framework | Knowledge-guided diffusion for pharmacophore mapping | Binding pose prediction, virtual screening |
| Cresset Field 3D-QSAR [23] | Methodology | 3D-QSAR using molecular field points | Activity prediction, lead optimization |
| DUD-E [21] | Database | Curated decoys for model validation | Virtual screening benchmarking |
| ZINC20 [25] [26] | Compound Database | Commercially available compounds for screening | Virtual screening library source |
These tools represent the technological infrastructure supporting advanced molecular alignment research. For instance, DiffPhore exemplifies the cutting-edge integration of artificial intelligence with traditional pharmacophore concepts, leveraging knowledge-guided diffusion frameworks for improved 3D ligand-pharmacophore mapping [25] [26]. Similarly, the Cresset Field 3D-QSAR method utilizes molecular field points derived from the Cresset XED force field as descriptors for QSAR models, enabling the capture of electrostatic and steric properties critical for biological activity [23].
The comparative analysis of pharmacophore mapping and common scaffold alignment reveals a complementary relationship rather than a competitive one between these foundational 3D-QSAR alignment methods. Common scaffold alignment demonstrates particular strength when working with congeneric series where a shared structural framework exists, enabling intuitive alignment and straightforward structure-activity relationship interpretation. Its performance excels in lead optimization contexts where incremental structural modifications are explored. Conversely, pharmacophore mapping offers superior versatility for scaffold hopping, target fishing, and cases with structurally diverse actives, as its feature-based abstraction captures essential interaction patterns independent of specific molecular frameworks. The emergence of AI-enhanced approaches like DiffPhore, which incorporates knowledge-guided diffusion models for pharmacophore mapping, further extends the capabilities of this paradigm by better handling conformational flexibility and directional constraints [25] [26]. The strategic selection between these methodologies should be guided by the structural diversity of the compound set, the specific drug discovery objective (lead identification vs. optimization), and the availability of structural information about the biological target.
In the field of computational drug design, Three-Dimensional Quantitative Structure-Activity Relationship (3D-QSAR) studies serve as a pivotal methodology for correlating the spatial characteristics of molecules with their biological activity. These approaches, including Comparative Molecular Field Analysis (CoMFA) and Comparative Molecular Similarity Indices Analysis (CoMSIA), rely on a fundamental prerequisite: the accurate spatial alignment of ligand molecules within a common coordinate system. The success and predictive power of the resulting models are profoundly influenced by the quality of these molecular overlays. For decades, researchers have faced the challenge of generating these alignments, often resorting to time-consuming manual methods that introduce subjectivity and require significant expert intervention. This comparison guide examines two automated solutions to this challenge: the Field-Based Similarity Searching (FBSS) method and the Steric and Electrostatic Alignment (SEAL) method, providing an objective analysis of their performance, underlying algorithms, and practical applications in contemporary drug discovery pipelines.
Theoretical Foundations: How FBSS and SEAL Approach the Alignment Problem
Core Principles of Field-Based Similarity Searching (FBSS)
The FBSS method operates on the principle that molecular recognition and binding are governed not by atomic positions per se, but by the molecular interaction fields surrounding the ligand. These fields represent the spatial distribution of properties critical to binding, such as steric bulk and electrostatic potential. The FBSS algorithm quantifies similarity by calculating the cosine coefficient between the field values of two molecules positioned within a three-dimensional grid, effectively measuring the congruence of their respective molecular landscapes [17]. This field-based approach offers a significant advantage: it can suggest non-obvious alignments that might be overlooked by manual methods focused on common substructures, thereby providing novel insights into structure-activity relationships.
Core Principles of Steric and Electrostatic Alignment (SEAL)
In contrast, the SEAL method employs a different strategy to achieve optimal molecular superposition. It utilizes a genetic algorithm to maximize an objective function that simultaneously optimizes the overlay of both steric and electrostatic potentials between molecules [17]. The objective function in SEAL is based on the formulation of similarity indices using Gaussian functions, which allow for the rapid evaluation of molecular similarity without the explicit use of a 3D grid. This method seeks a global solution to the alignment problem by efficiently exploring the conformational and orientational space, aiming to find the best mutual fit of the molecular fields of two or more structures.
Comparative Performance Analysis: FBSS vs. SEAL and Manual Methods
To objectively evaluate the practical utility of FBSS and SEAL, their performance must be examined against traditional manual alignments and against each other based on established benchmarks and validation datasets.
Statistical Performance in 3D-QSAR Modeling
The ultimate validation of any alignment method lies in the quality and predictive power of the 3D-QSAR models it produces. Research utilizing several literature datasets provides quantitative evidence for assessing these methods.
Table 1: Statistical Comparison of 3D-QSAR Models from Different Alignment Methods
| Alignment Method | Dataset(s) | QSAR Method | Predictive q² | Internal r² | Key Strengths |
|---|---|---|---|---|---|
| FBSS | Steroids, 5 other literature sets | CoMFA, CoMSIA | 0.6 - 0.8 (comparable to manual) [17] | 0.91 - 0.96 (comparable to manual) [17] | Fully automated; suggests non-obvious alignments; good starting point |
| SEAL | Information from general context | Maximizes Steric/Electrostatic Overlay | Not explicitly stated in search results | Not explicitly stated in search results | Optimizes steric/electrostatic fit simultaneously; Gaussian functions for speed |
| Manual (Reference) | Classic steroids, other benchmarks | CoMFA, CoMSIA | ~0.6 - 0.8 [17] | ~0.91 - 0.96 [17] | Leverages expert knowledge; can be time-consuming and subjective |
Experimental data confirms that FBSS-generated alignments produce 3D-QSAR models with predictive performance (q²) and internal consistency (r²) that are broadly comparable to those derived from manual alignments [17]. For instance, on a series of steroids and other literature datasets, FBSS enabled CoMFA and CoMSIA models with q² values in the range of 0.6-0.8 and r² values reaching 0.91-0.96, matching the standards set by painstaking manual methods [17]. This demonstrates that automation does not necessitate a sacrifice in model quality.
Key Differentiating Factors in Methodology and Application
While both are automated, fundamental differences in their algorithms lead to varying strengths.
Table 2: Methodological Comparison of FBSS and SEAL
| Feature | FBSS (Field-Based Similarity Searching) | SEAL (Steric and Electrostatic Alignment) |
|---|---|---|
| Primary Driver | Field similarity (Cosine coefficient) [17] | Similarity indices with Gaussian functions [17] |
| Algorithm Type | Field-based similarity calculation | Genetic algorithm optimizing an objective function [17] |
| Key Innovation | Application of database similarity searching to QSAR alignment | Simultaneous optimization of steric and electrostatic overlay [17] |
| Main Advantage | Can reveal non-obvious, field-based similarities | Efficiently finds a global optimum for field overlap |
| Typical Use Case | Initial automated screening and model generation | Finding an optimal alignment based on field congruence |
A critical insight from these comparisons is that FBSS serves not as a outright replacement for expert-driven manual alignment, but as a powerful complementary tool [17]. Its primary value lies in two scenarios: first, as an initial screening mechanism to rapidly determine if a dataset is amenable to 3D-QSAR analysis before investing significant manual effort; and second, as a source of novel alignment hypotheses that can inspire and guide subsequent, more detailed manual analyses [17].
Experimental Protocols for Method Validation
To ensure reproducibility and provide a clear framework for benchmarking, this section outlines the standard experimental procedures used to validate automated alignment methods like FBSS.
Standard Workflow for FBSS Validation in 3D-QSAR
The following diagram illustrates the sequential steps involved in a typical validation study for an automated alignment method.
1. Literature Dataset Curation: The process begins with the selection of well-characterized datasets from the published literature. These datasets must include experimental biological activity data (e.g., IC₅₀, Kᵢ) and ideally pre-defined training and test sets. Common benchmarks include the classic steroid dataset with binding affinity for corticosteroid-binding globulin and other sets relevant to targets like the farnesoid X receptor and opioid receptors [17] [27] [28].
2. Generate Molecular Conformations: Low-energy 3D conformations for each molecule in the dataset are generated. This often involves structure optimization using software like Sybyl-X [12].
3. Apply FBSS for Automated Alignment: The FBSS program is used to superpose all molecules in the dataset onto a chosen reference molecule. The alignment is driven by the optimization of the similarity of their molecular fields (steric and electrostatic) [17].
4. Perform 3D-QSAR (CoMFA/CoMSIA): The aligned molecule set is used as input for a 3D-QSAR analysis. The molecules are placed in a 3D grid, and their interaction fields are sampled. Partial Least Squares (PLS) regression is then used to derive the quantitative model linking the field values to the biological activity [17] [12].
5. Validate Model Statistically: The model is validated internally (e.g., using leave-one-out cross-validation to obtain q²) and externally by predicting the activity of the withheld test set compounds [27] [28]. A model with q² > 0.5 and a low standard error is generally considered predictive.
6. Compare with Manual Alignment: The final and crucial step is to compare the statistical performance and contour maps of the automated model with those from a model based on a manual alignment performed by domain experts [17].
Advanced Integration with Machine Learning
Recent advancements have begun to merge traditional 3D-QSAR with modern machine learning (ML) techniques. For example, after generating alignments and 3D molecular field descriptors, researchers can use algorithms like Random Forest (RF), Support Vector Machine (SVM), and Multilayer Perceptron (MLP) to build the predictive model instead of, or in comparison with, traditional PLS [29]. One study on estrogen receptor binding found that such ML-based 3D-QSAR models outperformed traditional 2D-QSAR models in terms of accuracy, sensitivity, and selectivity [29].
The Scientist's Toolkit: Essential Research Reagents and Software
Successful implementation of FBSS, SEAL, and related 3D-QSAR workflows requires a suite of specialized software tools and conceptual "reagents."
Table 3: Essential Resources for Field-Based Molecular Alignment and 3D-QSAR
| Tool/Resource | Type | Primary Function in Alignment/QSAR |
|---|---|---|
| FBSS Program | Software Module | Performs the field-based similarity calculations and automated molecular superposition [17]. |
| SEAL Algorithm | Software Algorithm | Maximizes the steric and electrostatic overlay between molecules using a genetic algorithm [17]. |
| Sybyl-X/Sybyl | Molecular Modeling Suite | Provides the environment for molecule construction, conformation optimization, and running CoMFA/CoMSIA analyses [12]. |
| CoMFA/CoMSIA | 3D-QSAR Methodology | Correlates molecular interaction fields (steric, electrostatic, etc.) with biological activity after alignment [17] [12]. |
| Molecular Field Descriptors | Computational Descriptor | Quantitative 3D grids of steric/electrostatic properties that drive FBSS alignment and form the variables for QSAR [17]. |
| Partial Least Squares (PLS) | Statistical Method | The regression technique used to relate the numerous field descriptors to biological activity in classical 3D-QSAR [17]. |
The drive toward automation in molecular alignment, exemplified by methods like FBSS and SEAL, represents a significant advancement in 3D-QSAR. The experimental evidence clearly demonstrates that these automated methods are no longer just conceptual shortcuts but are robust, reliable tools capable of producing predictive models that rival those derived from expert manual alignments. Their value in increasing throughput, reducing subjectivity, and generating novel structural insights is undeniable.
The future of this field lies in the continued refinement of these methods and their integration with other cutting-edge technologies. The application of more sophisticated machine learning algorithms for analyzing molecular field data is already showing promise [29]. Furthermore, the integration of alignment and 3D-QSAR within broader drug discovery workflows—including molecular docking, dynamics simulations, and advanced cheminformatics—will further cement their role as indispensable tools for the modern computational chemist [12]. As these tools become more accessible and user-friendly, their adoption will continue to grow, accelerating the rational design of novel therapeutic agents.
Shape-based molecular alignment is a foundational technique in modern computer-aided drug design that enables the comparison of molecules based on their three-dimensional steric and electrostatic properties rather than their two-dimensional topological structure. This approach is particularly valuable for identifying structurally diverse compounds that share similar biological activities, a process known as scaffold hopping. Unlike traditional 2D methods that rely on molecular graphs and substructure matching, 3D shape-based techniques can discover non-intuitive similarities between chemically distinct compounds by examining their volumetric characteristics and functional group orientations.
The application of shape-based alignment has revolutionized virtual screening by allowing researchers to identify potential drug candidates that would be missed by conventional similarity searches. This capability is especially crucial in early drug discovery when expanding chemical space exploration or designing novel patentable chemotypes is required. Tools like ROCS (Rapid Overlay of Chemical Structures) exemplify this methodology, using Gaussian-based shape representations and solid-body optimization to maximize volume overlap between molecules at speeds that make large-scale virtual screening practical [30]. The underlying principle posits that molecules adopting similar shapes and chemical feature distributions in 3D space are likely to interact with the same biological targets, even when their 2D structures appear quite different.
Fundamental Principles of Shape-Based Alignment
Molecular Shape Representation
Shape-based alignment tools employ different mathematical models to represent and compare molecular volumes:
Gaussian-Based Models: ROCS utilizes a Gaussian description of molecular shape that approximates hard-sphere volumes while enabling rapid similarity calculations. This approach represents molecules as collections of overlapping Gaussian functions centered on atomic positions, creating a smooth molecular surface that facilitates efficient volume overlap computation [30] [31]. The Gaussian method is parametrized to reproduce hard-sphere volumes while offering computational advantages for optimization.
Hard-Sphere Models: Alternative implementations like Schrödinger's Shape Screening represent structures as sets of hard atomic van der Waals spheres, with one sphere for each heavy atom and polar hydrogen. This approach computes overlap as the sum of pairwise atomic overlaps, ignoring intersections among three or more atoms to maximize calculation speed [32].
Chemical Feature Encoding ("Color" Force Fields)
Beyond pure shape, advanced shape-based methods incorporate chemical feature matching to improve biological relevance:
Feature-Based Scoring: ROCS extends shape matching with "color" force fields that encode chemical properties including hydrogen bond donors, acceptors, hydrophobic regions, and charged groups. These features are incorporated into the superposition scoring function, facilitating identification of compounds similar in both shape and key interaction capabilities [30] [33].
Pharmacophore Representation: Schrödinger's Shape Screening can alternatively represent structures as pharmacophore sites encoding hydrogen bond acceptors/donors, hydrophobic regions, ionizable functions, and aromatic rings, with each site represented by a 2Å hard sphere [32].
Similarity Scoring Metrics
The quantification of molecular similarity employs several specialized metrics:
Volume Overlap: The fundamental shape similarity measure compares the shared volume between two aligned structures relative to their total volume, typically expressed as
Shape Similarity = V_A∩B / V_A∪Bor normalized variants thereof [32].Composite Scoring: ROCS provides multiple scoring predicates including Tanimoto Combo (sum of shape and color Tanimoto scores), Fit Tversky, and Ref Tversky, allowing researchers to prioritize different aspects of molecular similarity for specific applications [33].
ElectroShape Similarity: Emerging approaches like ChemBounce implement Electron Shape similarity that considers both charge distribution and 3D shape properties, potentially offering enhanced biological activity preservation in scaffold hopping [34].
Scaffold Hopping Workflow Using Shape-Based Alignment
Table 1: Key Stages in Shape-Based Scaffold Hopping Workflow
| Stage | Key Activities | Tools & Techniques |
|---|---|---|
| Query Preparation | Select active compound; generate bioactive conformation; define core scaffold | Conformational analysis; scaffold identification algorithms (e.g., HierS) |
| Database Assembly | Curate screening collection; generate multi-conformer databases; apply filters | Rule-based fragmentation; scaffold libraries (e.g., ChEMBL-derived); diversity selection |
| Shape-Based Screening | Perform 3D alignment; compute shape similarity; apply chemical constraints | ROCS; Shape Screening; ElectroShape; triplet alignment algorithms |
| Hit Analysis & Validation | Examine top alignments; assess synthetic accessibility; prioritize candidates | Visual inspection; synthetic accessibility scoring (SAscore); property prediction |
| Experimental Verification | Synthesize selected analogs; determine biological activity; iterate design | Medicinal chemistry; biological assays; structure-activity relationship analysis |
The following workflow diagram illustrates the key stages and decision points in a typical shape-based scaffold hopping campaign:
Diagram 1: Scaffold Hopping Workflow via Shape-Based Alignment. This workflow outlines the systematic process for identifying novel scaffolds using 3D shape similarity, from initial query preparation through experimental verification.
Query Preparation and Scaffold Identification
The initial stage involves careful selection and preparation of the query molecule:
Template Selection: Choose a known active compound with well-characterized biological activity as the alignment template. High-affinity ligands with determined bioactive conformations (e.g., from crystal structures) yield optimal results [14].
Conformation Generation: For flexible molecules, multiple low-energy conformations should be generated to account for possible binding orientations. Tools like ConfGen or molecular dynamics simulations can produce biologically relevant conformers [32].
Scaffold Identification: Algorithms such as the HierS methodology systematically decompose molecules into ring systems, side chains, and linkers. Basis scaffolds are generated by removing all linkers and side chains, while superscaffolds retain linker connectivity, creating a hierarchy of structural components for replacement [34].
Database Assembly and Preparation
The screening database significantly impacts scaffold hopping success:
Diverse Scaffold Libraries: Curated scaffold collections, such as those derived from ChEMBL containing over 3 million unique synthesis-validated fragments, provide replacement candidates with high synthetic accessibility [34].
Multi-conformer Representation: Each database compound should be represented by multiple low-energy conformations to ensure shape complementarity can be properly evaluated despite molecular flexibility [32].
Drug-like Filtering: Application of property filters (molecular weight, logP, hydrogen bond donors/acceptors) and structural alerts helps prioritize compounds with favorable developability characteristics [34].
Shape-Based Screening and Alignment
The core computational phase performs 3D alignment and similarity assessment:
Rapid Triplet Alignment: Efficient methods identify numerous pairs of atom or pharmacophore triplets with similar geometries and local environments in query and database structures, superimposing molecules based on least-squares alignment of each triplet pair [32].
Volume Overlap Optimization: The initial alignments are refined by maximizing the volume overlap between molecules using either Gaussian-based [30] or hard-sphere [32] approaches.
Composite Similarity Scoring: Results are ranked using combined scores that balance shape complementarity and chemical feature overlap, such as Tanimoto Combo scores in ROCS [33] or pharmacophore-enriched similarity in Shape Screening [32].
Comparative Performance Analysis
Virtual Screening Enrichment
Table 2: Shape-Based Screening Performance Across Multiple Targets (Enrichment Factors at 1% of Database)
| Target | ROCS-Color | Schrödinger Shape Screening | SQW Method |
|---|---|---|---|
| CA | 31.4 | 32.5 | 6.3 |
| CDK2 | 18.2 | 19.5 | 9.1 |
| COX2 | 25.4 | 21.0 | 11.3 |
| DHFR | 38.6 | 80.8 | 46.3 |
| ER | 21.7 | 28.4 | 23.0 |
| HIV-PR | 12.5 | 16.9 | 5.9 |
| HIV-RT | 2.0 | 2.0 | 5.4 |
| Neuraminidase | 92.0 | 25.0 | 25.1 |
| PTP1B | 12.5 | 50.0 | 50.2 |
| Thrombin | 21.1 | 28.0 | 27.1 |
| TS | 6.5 | 61.3 | 48.5 |
| Average | 25.6 | 33.2 | 23.5 |
| Median | 21.1 | 28.0 | 23.0 |
Performance data from validated virtual screening benchmarks demonstrates significant variation between shape-based methods across different biological targets. Schrödinger's Shape Screening with pharmacophore representation shows particularly strong performance for DHFR, PTP1B, and TS targets, surpassing ROCS-color by 30-40% in average and median enrichments [32]. ROCS-color maintains robust performance across multiple targets, establishing it as a consistent performer, though the optimal tool appears target-dependent.
Technical Implementation Comparison
Table 3: Technical Characteristics of Shape-Based Alignment Tools
| Feature | ROCS/FastROCS | Schrödinger Shape Screening | ChemBounce |
|---|---|---|---|
| Shape Representation | Gaussian functions | Hard atomic spheres | Electron shape descriptors |
| Chemical Features | "Color" force fields | Atom typing or pharmacophore sites | ElectroShape similarity |
| Alignment Method | Solid-body optimization | Triplet alignment with refinement | Open-source algorithm |
| Speed | 600-800 conformers/second/CPU | ~600 conformers/second/CPU | 4s-21min per compound |
| GPU Acceleration | Yes (FastROCS) | Not specified | Not specified |
| Scaffold Hopping Focus | Established application | Primary capability | Explicit design purpose |
| Availability | Commercial | Commercial | Open-source |
| Special Capabilities | Composite queries; grid-based shapes | Multi-ligand superposition; excluded volumes | Synthetic accessibility focus |
The technical comparison reveals distinctive implementation strategies across platforms. ROCS employs Gaussian shape representation with solid-body optimization, while Shape Screening uses hard-sphere models with efficient triplet alignment [30] [32]. ChemBounce represents an open-source alternative specifically designed for scaffold hopping with integrated synthetic accessibility assessment [34]. FastROCS provides GPU acceleration for ultra-large library screening, processing millions of conformations per second [31].
Experimental Protocols for Validation
Standard Virtual Screening Protocol
To objectively evaluate shape-based tools, researchers have established standardized screening protocols:
Dataset Preparation: Compile a set of known actives for specific targets (e.g., CDK2, thrombin, HIV protease) and combine with decoy molecules (e.g., 25,000 MDDR compounds) representing drug-like chemical space [32].
Query Selection: For each target, select a single active compound with a determined bioactive conformation as the shape query.
Multi-conformer Database: Generate multiple low-energy conformations for all database compounds using tools like MacroModel or ConfGen [32].
Shape Screening Execution: Perform shape-based similarity search using each tool with consistent parameters. For comprehensive evaluation, include both shape-only and chemically-enabled modes.
Enrichment Calculation: Rank database compounds by similarity score and calculate enrichment factors (EF) at specific percentages (typically 1%) of the screened database:
EF = (Hitssampled / Nsampled) / (Hitstotal / Ntotal).
Scaffold Hopping Validation Metrics
Specific metrics for scaffold hopping success include:
Scaffold Diversity: Measure the structural diversity of identified hits using scaffold fingerprints or molecular frameworks to confirm genuine scaffold hops beyond simple analog finding.
Tanimoto Similarity: Calculate 2D structural similarity between query and hits to ensure identified compounds represent significant structural departures (typically <0.3 Tanimoto similarity for true scaffold hops).
Shape Similarity Thresholds: Apply appropriate similarity cutoffs (e.g., Tanimoto Combo >1.2 in ROCS) to balance novelty and activity retention [34].
Synthetic Accessibility: Assess the synthetic tractability of proposed scaffold hops using metrics like SAscore to prioritize feasible candidates for synthesis [34].
Table 4: Key Resources for Shape-Based Scaffold Hopping
| Resource Category | Specific Examples | Function in Scaffold Hopping |
|---|---|---|
| Software Tools | ROCS/FastROCS; Schrödinger Shape Screening; ChemBounce | Perform 3D shape-based alignment and similarity calculations |
| Scaffold Libraries | ChEMBL-derived fragments; proprietary corporate collections | Provide diverse replacement scaffolds with known synthesis routes |
| Conformation Generators | ConfGen; MacroModel; RDKit | Generate biologically relevant 3D conformations for screening |
| Chemical Databases | Enamine REAL Space; ZINC; commercial screening collections | Source compounds for virtual screening or purchase |
| Synthetic Planning Tools | SAscore; retrosynthesis software; PReal | Assess and plan synthesis of proposed scaffold hops |
| Validation Assays | Binding assays; functional cellular assays; structural biology | Confirm retained activity of scaffold-hopped compounds |
Shape-based alignment represents a powerful strategy for scaffold hopping, complementing other computational approaches in the medicinal chemist's toolkit. The comparative analysis demonstrates that while ROCS establishes a strong performance baseline with its Gaussian shape representation and chemical feature matching, alternative implementations like Schrödinger's Shape Screening with pharmacophore encoding can achieve superior enrichment for specific targets. The emergence of open-source platforms like ChemBounce expands accessibility while incorporating modern considerations like synthetic accessibility and electron shape similarity.
Future developments in shape-based scaffold hopping will likely focus on integrating artificial intelligence for enhanced similarity assessment, leveraging ultra-large virtual libraries enabled by GPU acceleration [31], and combining shape-based with structure-based methods in unified workflows. As shape-based alignment continues to evolve, its role in accelerating the discovery of novel bioactive chemotypes through efficient scaffold hopping appears increasingly secured, providing valuable intellectual property opportunities and chemical starting points for drug discovery campaigns.
Quantitative Structure-Activity Relationship (QSAR) modeling represents a cornerstone of modern computational drug discovery, enabling researchers to predict biological activity from chemical structure. Traditional three-dimensional QSAR (3D-QSAR) techniques, such as Comparative Molecular Field Analysis (CoMFA), rely heavily on the spatial superimposition of ligand molecules, requiring them to be aligned in a consistent frame of reference based on their putative binding mode [8]. This alignment-dependent paradigm, while powerful, introduces substantial methodological challenges, particularly when dealing with structurally diverse compounds that lack a common molecular scaffold [35].
The pursuit of alignment-independent methodologies has emerged as a critical research direction to overcome these limitations. Among the most significant advancements in this domain are Grid-Independent Descriptors (GRIND), which fundamentally reimagine how molecular interaction fields are captured and quantified [36]. This guide provides a comprehensive comparison of these alignment-independent techniques against traditional methods, examining their underlying principles, experimental protocols, and performance across diverse drug discovery applications.
The GRIND Descriptor Framework
Fundamental Principles and Workflow
GRIND descriptors are alignment-independent molecular descriptors derived from Molecular Interaction Fields (MIFs) [36]. Unlike conventional 3D-QSAR approaches that require meticulous molecular superimposition, GRIND captures relevant molecular characteristics without spatial alignment, making them particularly valuable for handling structurally diverse compound sets [37].
The GRIND calculation process involves three fundamental steps:
MIF Calculation: A set of molecular interaction fields is computed for each molecule using various probes (e.g., DRY for hydrophobic interactions, N1 for hydrogen bond acceptor, O for hydrogen bond donor) [36] [37]. These MIFs represent the interaction energies between the molecule and specific probes at numerous points around the molecular structure.
Node Filtering: The MIFs are filtered to extract the most relevant regions, resulting in "final nodes" that represent favorable probe-target interaction regions [36].
Descriptor Encoding: The filtered MIFs are encoded into GRIND variables by computing the product of interaction energies for each pair of nodes and sorting these products according to the distance between nodes [36]. The highest product value in each distance category is stored, creating a distance-based correlation known as a correlogram [8].
Enhanced Shape Descriptors
A significant advancement in the GRIND methodology came with the incorporation of molecular shape descriptors. The original GRIND approach recognized limitations in adequately describing ligand shape, which often plays a crucial role in receptor binding [38]. This led to the development of enhanced descriptors that incorporate molecular surface curvature measurements, providing a more comprehensive characterization of molecular morphology and enabling the identification of both favorable and unfavorable shape complementarity in ligand-receptor interactions [38].
Table: Key Probes Used in GRIND Descriptor Generation
| Probe Type | Representation | Primary Application |
|---|---|---|
| DRY | Hydrophobic interactions | Mapping hydrophobic contact regions |
| N1 | Amide nitrogen (H-bond acceptor) | Identifying hydrogen bond acceptor sites |
| O | Carbonyl oxygen (H-bond donor) | Identifying hydrogen bond donor sites |
| TIP | Molecular shape/bulk | Characterizing steric properties |
Experimental Protocols and Implementation
Standard GRIND-Based 3D-QSAR Workflow
The implementation of GRIND-based 3D-QSAR follows a structured experimental pathway with specific requirements at each stage:
Dataset Preparation and Conformational Analysis
- Compound Selection: Curate a structurally diverse set of compounds with consistent biological activity data (e.g., IC50, EC50 values) [37] [39].
- 3D Structure Generation: Convert 2D structures to 3D coordinates using programs such as CORINA or Omega [36].
- Conformational Sampling: Perform thorough conformational analysis to identify the lowest energy conformation for each compound. Studies indicate that using the lowest energy conformer typically yields the most reliable models, though some investigations explore the impact of multiple conformations [36] [39].
GRIND Descriptor Calculation
- Software Implementation: Utilize specialized software such as ALMOND for descriptor generation [36].
- MIF Computation: Calculate molecular interaction fields using selected probes (typically DRY, N1, O, and TIP) [37].
- Distance Binning: Apply a distance-based encoding scheme with discrete bins (e.g., 0.5 Å increments from 0.33 to 25.83 Å) [39].
Model Development and Validation
- Data Splitting: Divide the dataset into training and test sets using activity-stratified partitioning to ensure representative distribution [39] [23].
- Statistical Analysis: Employ Partial Least Squares (PLS) regression or machine learning algorithms (e.g., Random Forest, SVM) to correlate descriptors with biological activity [39] [23].
- Model Validation: Apply both internal (cross-validation, leave-one-out) and external (test set prediction) validation methods [37] [35]. Key validation metrics include q² for internal predictive ability and r² for external validation.
The following workflow diagram illustrates the complete GRIND-based 3D-QSAR process:
Representative Case Study Protocol
A study on S1P1 receptor agonists exemplifies a well-executed GRIND implementation [37]:
- Dataset: 62 S1P1 receptor agonists based on a 2-imino-thiazolidin-4-one scaffold with experimentally determined EC50 values.
- Structure Preparation: 3D structures generated using HyperChem software (version 8.0.8) with geometry optimization performed via MM+ force field followed by semi-empirical AM1 algorithm.
- Descriptor Calculation: GRIND descriptors computed using ALMOND software with standard probes (DRY, N1, O, TIP).
- Model Construction: PLS regression applied to correlate GRIND descriptors with pEC50 values.
- Validation: Both internal (cross-validation, r²acc = 0.93) and external (test set prediction, r² = 0.75) validation performed.
- Virtual Screening: Resulting model applied to screen PubChem database, identifying four potential S1P1 receptor agonists with high predicted potency and selectivity.
Comparative Performance Analysis
Quantitative Comparison of Methodologies
Direct comparison studies reveal distinct performance characteristics between alignment-independent and traditional 3D-QSAR approaches:
Table: Performance Comparison of 3D-QSAR Methodologies
| Methodology | Alignment Requirement | Structural Flexibility Handling | Typical q² Values | Best Application Context |
|---|---|---|---|---|
| GRIND | Not required | Excellent | 0.69 - 0.82 [36] [37] | Structurally diverse compounds, congeneric series |
| CoMFA | Critical | Moderate | 0.60 - 0.75 [8] | Congeneric series with common scaffold |
| CoMSIA | Critical | Moderate | 0.65 - 0.78 [8] | Cases requiring hydrogen bonding description |
| Quantum 3D-QSAR | Critical (optimized) | Challenging | 0.79+ [35] | Targets where electronic effects dominate |
Application Case Studies Across Target Classes
Enzyme Targets: OSC and SHC Inhibitors
A rigorous evaluation of GRIND capabilities examined its performance in predicting inhibitory activity for two similar enzymes, oxidosqualene cyclase (OSC) and squalenehopene cyclase (SHC) [36]. The study utilized 28 non-terpenoid inhibitors and demonstrated that GRIND-based models could reliably predict both inhibitory activities despite the similar active site architecture of the two enzymes. The resulting models showed excellent predictive performance with the methodology correctly identifying differential structural requirements for inhibition of the two similar enzyme targets.
Ion Channel Targets: hERG Blocker Prediction
Cardiotoxicity prediction through hERG channel blocking represents a crucial application in drug safety assessment. A GRIND-based 3D-QSAR study on hERG K+ channel blockers addressed this challenging endpoint [36]. The investigation yielded a robust three-latent-variable model (r² = 0.93, qLOO² = 0.69) capable of identifying critical structural features associated with hERG blockade. This demonstrates GRIND's utility in predicting complex physicochemical interactions that govern off-target pharmacological effects.
GPCR Targets: S1P1 Receptor Agonists
Research on Sphingosine 1-phosphate type 1 (S1P1) receptor agonists highlighted GRIND's capability to model receptor selectivity [37]. The study developed predictive models for both S1P1 and S1P3 receptor agonism, enabling the identification of selective S1P1 receptor agonists with reduced potential for side effects mediated by S1P3 receptor activation. The resulting model achieved impressive internal (r²acc = 0.93) and external (r² = 0.75) predictivity, leading to the identification of four novel potential S1P1 selective agonists through virtual screening.
Unique Applications: Ice Recrystallization Inhibitors
Beyond conventional drug targets, GRIND has demonstrated utility in specialized applications such as cryopreservation. A study aimed at discovering novel ice recrystallization inhibitors (IRIs) utilized GRIND descriptors to model this unusual property [39]. The research employed a diverse set of 124 carbohydrate-based molecules, with GRIND descriptors calculated from quantum mechanically derived electrostatic potentials and molecular surface curvatures. The resulting classification model successfully identified 82% of novel active compounds in experimental validation, showcasing the method's adaptability to non-pharmacological endpoints.
Research Reagent Solutions
Table: Essential Computational Tools for GRIND-Based 3D-QSAR
| Tool Category | Specific Software/Resource | Primary Function | Application Note |
|---|---|---|---|
| Descriptor Generation | ALMOND [36] | GRIND descriptor calculation | Industry-standard for alignment-independent 3D-QSAR |
| Structure Preparation | CORINA, Omega [36] | 3D structure generation | Convert 2D structures to 3D coordinates for analysis |
| Quantum Chemistry | Gaussian '09 [39] | Electronic structure calculation | Provides accurate electrostatic potentials for GRIND |
| Molecular Modeling | HyperChem [37] | Molecular mechanics/dynamics | Force field-based geometry optimization |
| Statistical Analysis | PLS Toolboxes, R packages [39] | Multivariate data analysis | Correlation of descriptors with biological activities |
| Virtual Screening | PubChem Database [37] | Compound library sourcing | Source of novel structures for predictive screening |
Alignment-independent 3D-QSAR techniques, particularly those utilizing GRIND descriptors, represent a significant methodological advancement in computational drug discovery. The evidence from comparative studies indicates that these methods achieve predictive performance comparable to traditional alignment-dependent approaches while offering substantial advantages in handling structural diversity and eliminating alignment subjectivity [36] [37] [39].
GRIND-based methodologies have demonstrated exceptional versatility across multiple target classes, including enzymes, ion channels, and GPCRs, while also proving adaptable to specialized applications beyond conventional drug discovery [36] [37] [39]. The incorporation of molecular shape descriptors has further enhanced their capability to model complex steric requirements for receptor binding [38].
As drug discovery increasingly focuses on challenging targets and structurally diverse compound libraries, alignment-independent techniques like GRIND are poised to play an expanding role in the computational chemist's toolkit. Their integration with advanced machine learning algorithms and quantum chemical descriptors represents a promising direction for further enhancing predictive accuracy and mechanistic interpretability in 3D-QSAR modeling [35] [23].
Within the field of computer-aided drug design, Three-Dimensional Quantitative Structure-Activity Relationship (3D-QSAR) modeling serves as a pivotal technique for correlating the biological activity of compounds with their three-dimensional structural and electronic properties. A foundational yet often unstated prerequisite for many 3D-QSAR methods, such as Comparative Molecular Field Analysis (CoMFA) and Comparative Molecular Similarity Indices Analysis (CoMSIA), is the requirement for a valid molecular alignment. The choice of alignment strategy—whether manual, automated, or receptor-based—can profoundly influence the predictive power and interpretability of the resulting model. This case study provides a comparative analysis of two historically significant datasets: the classic steroid benchmark and contemporary Monoamine Oxidase B (MAO-B) inhibitors. The steroid dataset, once the gold standard for method validation, now illustrates critical pitfalls, while MAO-B inhibitor studies exemplify the modern, multi-faceted approaches required for robust 3D-QSAR in drug discovery. This analysis is framed within the broader thesis that the evolution of alignment methods reflects a growing recognition of dataset-specific limitations and a shift towards integrative, biologically-relevant modeling strategies.
Dataset Profiles and Historical Context
Table 1: Core Characteristics of the Steroid and MAO-B Inhibitor Datasets
| Feature | Steroid Dataset | MAO-B Inhibitor Datasets |
|---|---|---|
| Primary Application | Benchmarking 3D-QSAR methods (historically) | Drug discovery for neurodegenerative diseases (e.g., Parkinson's) |
| Key Molecular Target | Corticosteroid Binding Globulin (CBG) | Monoamine Oxidase B (MAO-B) Enzyme |
| Notable Dataset Size | 31 compounds (classic set) | Varies (e.g., 126 to over 450 compounds in modern studies) |
| Structural Nature | Structurally congeneric and rigid | Chemically diverse, including coumarins, chromones, chalcones, and benzothiazoles |
| Inherent Flexibility | Low | Moderate to High |
The Steroid Benchmark Dataset
For approximately two decades, a set of 31 steroids binding to corticosteroid-binding globulin (CBG) was the standard benchmark for evaluating 3D-QSAR methods [40]. Its popularity stemmed from the structural congenericity and relative rigidity of steroid molecules, which simplified the molecular alignment process. This perceived simplicity made it an attractive test case for demonstrating the statistical performance of new 3D-QSAR descriptors and methodologies.
MAO-B Inhibitor Datasets
In contrast, MAO-B inhibitor datasets are driven by clear therapeutic objectives. MAO-B is a well-characterized flavoprotein enzyme targeted for the treatment of Parkinson's and Alzheimer's diseases [41] [42]. Modern datasets comprise hundreds of chemically diverse compounds, including derivatives of coumarins, chromones, chalcones, and benzothiazole-2-carboxamides [43] [44] [12]. This structural heterogeneity presents a significant challenge for alignment but more accurately represents the real-world drug discovery environment.
Critical Analysis of Methodological Approaches
The Steroid Dataset: A Cautionary Tale
A seminal 2009 study revealed a critical flaw in the steroid dataset and other popular "benchmarks" [40]. Researchers demonstrated that models with comparable statistical performance could be built using extremely simple descriptors, including binary occupancy indicators that neglected all chemical information. Astonishingly, for most datasets examined, models required descriptors from fewer than twelve—and in one case, just one—key atomic positions to perform nearly as well as models using sophisticated 3D descriptors like those in CoMFA.
This finding suggests that for these specific datasets, the high predictive power was not necessarily due to the method's ability to capture nuanced 3D physicochemical fields, but rather its capacity to identify a few spatial regions where simply "filling space" correlated with enhanced activity. The authors concluded that these datasets, including the steroid set, cannot reliably distinguish the merits of different 3D-QSAR descriptors and advocated for the use of simulated data for benchmarking purposes [40].
Modern 3D-QSAR Workflows for MAO-B Inhibitors
Contemporary studies on MAO-B inhibitors have moved beyond reliance on a single alignment method, adopting integrated workflows that combine multiple computational techniques to enhance reliability.
Table 2: Comparison of Alignment and Modeling Methodologies
| Methodology | Application in Steroid Studies | Application in MAO-B Inhibitor Studies | Key Advantage | Key Limitation |
|---|---|---|---|---|
| Manual Alignment | Common, based on common steroid core [40] | Less common due to high structural diversity | Intuitive, leverages expert knowledge | Subjective, time-consuming, difficult for diverse sets |
| Automated Field-Based Alignment (e.g., FBSS) | Shown to be comparable to manual alignment [17] | Used as an initial screening tool | Objective, reproducible, fast | Alignments may be dominated by pharmacologically irrelevant features |
| Docking-Based Alignment | Not typically used | Standard practice (e.g., with MAO-B crystal structure) [42] [12] | Provides biologically relevant pose within protein active site | Dependent on accuracy of docking scoring function |
| Alignment-Independent 3D-QSDAR | Not applied in reviewed studies | Successful application for androgen receptor binders, suggesting utility for MAO-B [10] | Bypasses alignment entirely, uses internal coordinates | Relies on different descriptor types (e.g., NMR chemical shifts) |
A typical modern protocol, as applied to unsaturated ketone derivatives and 6-hydroxybenzothiazole-2-carboxamides, involves a multi-step process [42] [12]:
- Docking-Based Alignment: Molecules are docked into the crystal structure of the MAO-B enzyme, and the resulting poses are used for alignment in subsequent CoMFA/CoMSIA studies.
- 3D-QSAR Model Building: Robust CoMSIA models are developed, often revealing the importance of steric, electrostatic, and hydrogen-bond acceptor fields for activity [42].
- Validation and Design: The model is used to predict the activity of novel designed compounds.
- Molecular Dynamics (MD) Simulation: The stability of top-ranked compounds bound to MAO-B is verified through MD simulations (e.g., 100 ns), assessing metrics like Root Mean Square Deviation (RMSD) [41] [12].
- Binding Affinity Calculation: Methods like MM-GBSA are used to calculate binding free energies, confirming interactions with key residues such as Tyr435, Tyr326, and Cys172 [42] [44].
This workflow synergistically combines ligand- and structure-based approaches, mitigating the limitations inherent in any single method.
Experimental Protocols and Data Comparison
Protocol: Building a Traditional 3D-QSAR Model (Steroid-like)
- Data Curation: A set of compounds with measured biological activity (e.g., IC50, Ki) is assembled.
- Conformational Sampling and Alignment: A low-energy conformation for each molecule is generated. For congeneric series, a common scaffold is identified and used for rigid body alignment.
- Descriptor Calculation: The aligned molecules are placed in a 3D grid, and steric (Lennard-Jones) and electrostatic (Coulombic) field energies are calculated at each grid point using a probe atom.
- Statistical Analysis: Partial Least Squares (PLS) regression is used to correlate the field descriptors with the biological activity, generating a predictive model. Key metrics include q² (cross-validated R²) and r² (conventional R²).
Protocol: Building an Integrated MAO-B Inhibitor Model
- Data Curation & Preparation: A diverse set of MAO-B inhibitors with consistent biological data is curated. Structures are prepared (e.g., energy minimization) [12].
- Docking-Based Alignment: The crystal structure of MAO-B (e.g., PDB: 2V5Z) is prepared. All compounds are docked into the active site, and the top-scoring pose for each is used for alignment [42].
- 3D-QSAR and Validation: CoMFA/CoMSIA models are built and validated. The model is used to predict novel compounds, considering pharmacokinetic (ADMET) properties [42].
- Molecular Dynamics Simulation: The complex of a promising inhibitor with MAO-B is solvated in an explicit water box and subjected to a 100 ns MD simulation. RMSD, RMSF, and protein-ligand contacts are analyzed to assess stability [41] [42].
- Binding Free Energy Calculation: Using frames from the stable MD trajectory, the binding free energy is computed via the MM-GBSA method, and per-residue energy decomposition is performed to identify key interactions [42].
Comparative Performance Data
Table 3: Representative Quantitative Outputs from Case Studies
| Study Dataset | QSAR Model Type | Key Statistical Metrics | Key Findings/Outcomes |
|---|---|---|---|
| Steroids | Simple Occupancy Descriptors [40] | Near-equivalent performance to CoMFA | Models required <12 atomic positions; questions validity of benchmark. |
| 6-hydroxybenzothiazole-2-carboxamides [12] | COMSIA | q² = 0.569, r² = 0.915 | Successfully guided design of compound 31.j3 with high predicted activity and stable MD profile (RMSD 1.0-2.0 Å). |
| Unsaturated Ketone Derivatives [42] | CoMFA & COMSIA | Not explicitly reported | Identified key fields; designed compound T1 with superior predicted binding affinity (ΔG = -409.5 kJ/mol) vs. original lead. |
| Diverse MAO-B Inhibitors (126 compounds) [44] | Pharmacophore-based 3D-QSAR | R² = 0.900, Q² = 0.774 | Model highlighted two H-bond acceptors, one hydrophobic, and one aromatic ring as critical for activity. |
Visualization of Workflows
The Scientist's Toolkit: Essential Research Reagents and Solutions
Table 4: Key Computational Tools for 3D-QSAR and Alignment
| Tool / Resource | Function / Application | Relevance to Alignment |
|---|---|---|
| Sybyl-X Software | Molecular modeling and analysis; contains modules for CoMFA and CoMSIA. | Provides tools for both manual and automated (e.g., field-fit) alignment of molecules. |
| Protein Data Bank (PDB) ID: 2V5Z | Crystal structure of human Monoamine Oxidase B (MAO-B). | Enables structure-based alignment; ligands can be docked and aligned within this active site. |
| FBSS (Field-Based Similarity Searching) | Program for automated generation of molecular alignments based on field similarity. | Offers an objective alternative to manual alignment, shown to produce models comparable to manual methods [17]. |
| 3D-QSDAR Methodology | Alignment-independent 3D-QSAR technique using descriptors from internal coordinates. | Circumvents the alignment problem entirely, useful for large, diverse datasets [10]. |
| GROMACS / AMBER | Software for Molecular Dynamics (MD) simulations. | Validates the stability of ligand poses obtained from docking-based alignment in a simulated biological environment. |
| Python (RDKit, Scikit-learn) | Programming environment for cheminformatics and machine learning. | Facilitates the calculation of 2D descriptors and the development of machine learning-QSAR models as a complementary approach [45]. |
This comparative analysis underscores a critical evolution in 3D-QSAR practices, directly tied to the choice and application of molecular alignment methods. The historical reliance on small, congeneric, and rigid datasets like the steroids, while convenient, obscured a significant methodological vulnerability: the inability of such datasets to truly validate the predictive power of 3D physicochemical fields, leading to potentially spurious correlations. The modern paradigm, exemplified by MAO-B inhibitor research, embraces complexity and mitigates alignment subjectivity through integrative workflows. By combining docking-based alignment with biologically relevant protein structures, and further validating models with MD simulations and binding energy calculations, researchers can construct more robust and predictive 3D-QSAR models. The broader thesis is clear: the field has matured from seeking universal benchmarks to adopting context-driven, multi-technique strategies that prioritize biological relevance over computational convenience, ensuring that 3D-QSAR remains a powerful tool in rational drug design.
Overcoming Alignment Challenges: Strategies for Robust and Predictive 3D-QSAR Models
In three-dimensional quantitative structure-activity relationship (3D-QSAR) studies, molecular flexibility presents a fundamental challenge for accurate model prediction. The core of this challenge lies in selecting appropriate molecular conformations to represent each compound in the dataset. The "global minimum dilemma" refers to the longstanding assumption in molecular modeling that the most biologically relevant conformation corresponds to the global minimum energy state of the isolated molecule. However, crystallographic evidence demonstrates that crystalline packing forces can stabilize conformers with energies up to 20 kJ mol⁻¹ above the global minimum, with these higher-energy conformers often being more extended to allow for greater intermolecular stabilization [46]. This discrepancy between isolated molecule energetics and biologically relevant conformations necessitates sophisticated approaches to conformer selection in 3D-QSAR workflows, particularly as the field advances toward more flexible drug-like molecules.
Theoretical Framework: Quantifying Molecular Flexibility
Traditional and Emerging Flexibility Metrics
Accurately quantifying molecular flexibility is essential for understanding its impact on conformer selection. Traditional metrics have included simple descriptors such as rotatable bond counts, while more sophisticated approaches have emerged to provide continuous descriptions of conformational space.
- Rotatable Bond Count: The most common definition includes single, non-ring bonds connecting two atoms each possessing at least one non-terminal substituent. While easily computed, this descriptor provides only a coarse-grained, integer-value view of conformational space and fails to account for varying degrees of rotational freedom in different bond types [47].
- Kier Flexibility Index (ϕ): Developed as a continuous descriptor derived from molecular topology, ϕ quantifies how structural features (branching, cyclization, heteroatoms) decrease flexibility from an idealized infinite carbon chain reference. While an improvement over rotatable bond counts, the Kier index cannot distinguish stereo- or regioisomers [47].
- Torsion Angular Bin Strings (TABS) and nTABS: This emerging approach discretizes the conformational space by representing each conformer as a vector of binned dihedral angles (TABS). The number of possible distinct TABS (nTABS) for a molecule provides an estimate of conformational ensemble size that accounts for variations in torsion state multiplicities and can differentiate between isomers [47].
Table 1: Comparison of Molecular Flexibility Metrics
| Metric | Description | Advantages | Limitations |
|---|---|---|---|
| Rotatable Bond Count | Number of bonds meeting specific rotatability criteria | Simple, fast to compute | Coarse-grained; ignores bond-specific torsion profiles |
| Kier ϕ Index | Topological descriptor based on κ shape indices | Continuous value; accounts for branching and cyclization | Cannot distinguish isomers; derived solely from 2D structure |
| nTABS | Product of possible torsion states for each rotatable bond | Accounts for bond-specific torsion multiplicities; distinguishes isomers | Requires reference torsion data; may overcount correlated torsions |
The Conformer Selection Landscape
The selection of molecular conformations for 3D-QSAR modeling spans a spectrum from single-conformer to ensemble-based approaches, each with distinct implications for addressing the global minimum dilemma:
- Single Conformer Approaches: Traditional 3D-QSAR methods typically utilize one conformation per molecule, often selected based on lowest energy criteria or structural similarity to a known active compound. These methods face significant limitations when biological activity arises from multiple conformations or when crystal packing forces stabilize higher-energy states [46].
- Multi-Instance Learning: This approach incorporates multiple conformations during model training, allowing algorithms to automatically identify plausible bioactive conformations without predefined selection rules. Studies demonstrate that MI-QSAR frequently outperforms single-instance approaches, particularly for flexible molecules with complex structure-activity relationships [48].
- Energy-Structure Integration: Evidence suggests that considering both conformational energy and molecular surface area improves predictions of crystalline conformations. A parameterized pseudo-energy term incorporating surface area leads to dramatic improvements in predicting conformations adopted in crystal structures [46].
Comparative Analysis of Computational Strategies
Methodological Implementations and Performance
Recent computational advancements have produced diverse strategies for handling molecular flexibility in drug discovery applications, with significant implications for 3D-QSAR conformer selection.
Table 2: Comparison of Computational Methods Addressing Molecular Flexibility
| Method | Approach | Conformer Selection Strategy | Key Advantages |
|---|---|---|---|
| SCAGE [49] | Self-conformation-aware graph transformer | Multiscale conformational learning from lowest-energy and varied-energy conformations | Integrates 3D information directly into architecture; functional group annotation |
| MI-QSAR [48] | Multi-instance learning with conventional and deep learning algorithms | Uses multiple conformations per molecule; automatically identifies bioactive conformations | Outperforms single-instance QSAR in numerous cases; no predefined conformation selection needed |
| Py-CoMSIA [16] | Open-source 3D-QSAR implementation | Typically relies on user-defined conformer selection and alignment | Gaussian function eliminates sharp cutoffs; less sensitive to alignment than CoMFA |
| Molecular Dynamics | Sampling of thermodynamic ensemble | Explicit simulation of conformational transitions under specified conditions | Accounts for solvation and temperature effects; provides kinetic information |
| CSP Methods [46] | Crystal structure prediction with DFT-D | Assesses crystallizability based on energy and surface area | Quantifies packing-induced strain; identifies stable crystalline conformations |
The Global Minimum in Experimental and Computational Contexts
The assumption that the global minimum energy conformation represents the biologically relevant state requires critical examination in light of experimental evidence:
- Crystallographic Validation: Analysis of pharmaceutical-like molecules reveals that crystal structures frequently adopt conformations with energies significantly above the global minimum (ΔEconf). The maximum observed strain energy reaches 20 kJ mol⁻¹, demonstrating the substantial distorting power of crystal packing forces [46].
- Surface Area Relationship: Higher-energy conformers observed in crystal structures tend to be more extended than their lower-energy counterparts, providing greater molecular surface area for intermolecular interactions. This suggests that crystallizability depends on both energy and surface area [46].
- Implications for 3D-QSAR: These findings challenge the automatic selection of global minimum conformations for 3D-QSAR studies, particularly when modeling binding to protein active sites where packing forces analogous to crystallization may stabilize non-minimum states.
Experimental Protocols and Workflow Implementation
Standardized Conformer Selection Methodology
To ensure reproducible and biologically relevant conformer selection in 3D-QSAR studies, the following experimental protocol is recommended:
Conformer Generation: Utilize distance geometry algorithms (e.g., ETKDGv3 in RDKit) with torsion preferences derived from crystallographic databases to generate initial conformational ensembles [47]. For each molecule, generate a minimum of 50-100 conformers, with increased sampling for molecules with higher flexibility (nTABS > 100).
Energy Evaluation and Filtering: Optimize generated conformers using molecular mechanics (MMFF) or density functional theory (DFT-D) methods. Retain conformers within a defined energy window (typically 10-20 kJ mol⁻¹ above the global minimum) to include potentially relevant higher-energy states [46].
Representative Selection: Apply clustering algorithms (RMSD or TFD-based) to identify structurally diverse representatives. For multi-instance QSAR, include representatives from major clusters; for single-conformer methods, select the centroid of the largest cluster or the conformation most similar to known active compounds [48].
Alignment for 3D-QSAR: Superimpose selected conformers using common substructures or pharmacophore points. For CoMSIA implementations, ensure consistent alignment rules across the entire dataset [16].
Multi-Instance Learning Implementation
For MI-QSAR applications, implement the following specialized protocol:
Conformer Ensemble Preparation: Generate a minimum of 10-20 conformers per compound using the standardized protocol above. Energy-based filtering may employ a wider threshold (up to 50 kJ mol⁻¹) to ensure coverage of potential bioactive states [48].
Descriptor Calculation: Compute 3D molecular descriptors (e.g., CoMSIA fields, molecular shape, electrostatic potentials) for each conformation in the ensemble [16].
Model Training with Instance Bagging: Treat all conformations of a single molecule as instances within a "bag" labeled with the molecule's experimental activity. Implement multi-instance algorithms that can handle this representation during training [48].
Bioactive Conformer Identification: Utilize attention mechanisms or instance weighting in deep learning architectures to automatically identify which conformations contribute most to activity predictions [49].
The following workflow diagram illustrates the decision process for conformer selection strategies in 3D-QSAR studies:
Research Reagent Solutions: Essential Computational Tools
Successful implementation of conformer selection strategies requires specialized computational tools and resources. The following table details essential "research reagents" for addressing molecular flexibility in 3D-QSAR studies.
Table 3: Essential Computational Tools for Conformer Selection and Analysis
| Tool/Resource | Type | Primary Function | Application in Conformer Selection |
|---|---|---|---|
| RDKit [47] | Open-source cheminformatics library | Molecular informatics and machine learning | Conformer generation (ETKDGv3), rotatable bond identification, descriptor calculation |
| CRYSTAL09 [46] | Quantum chemistry software | Periodic DFT calculations for molecular crystals | High-accuracy conformational energy calculations; crystal structure optimization |
| Py-CoMSIA [16] | Open-source Python library | 3D-QSAR modeling with CoMSIA method | Implementation of Gaussian-based similarity fields; less sensitive to alignment issues |
| ChEMBL Database [50] | Bioactivity database | Curated bioactivity data for drug discovery | Source of experimental activity data for QSAR model building and validation |
| Cambridge Structural Database [47] | Crystallographic database | Experimental small-molecule crystal structures | Source of torsion angle distributions for conformer generation parameterization |
| MacroModel [46] | Molecular modeling software | Molecular mechanics and conformational analysis | Low-mode conformational search (LMCS) for comprehensive conformer generation |
The challenge of molecular flexibility in 3D-QSAR represents both a significant obstacle and opportunity for advancing drug discovery methodologies. The global minimum dilemma necessitates a paradigm shift from single-conformer approaches toward ensemble-based strategies that acknowledge the complex interplay between conformational energetics and biological environment. Emerging methodologies including multi-instance learning, deep learning architectures with integrated conformational awareness (SCAGE), and advanced flexibility metrics (nTABS) offer promising avenues for more accurate bioactivity prediction. Future developments will likely focus on dynamic conformational sampling under biologically relevant conditions, integration of protein flexibility, and automated identification of bioactive conformations through explainable AI approaches. As these methodologies mature, they will progressively resolve the longstanding challenges posed by molecular flexibility in quantitative structure-activity relationship modeling.
The predictive power of Three-Dimensional Quantitative Structure-Activity Relationship (3D-QSAR) models is fundamentally dependent on the spatial alignment of the molecules under study. This process becomes particularly challenging when dealing with structurally diverse datasets containing scaffold hops—compounds with different core structures (backbones) that share similar biological activity. Effective handling of these alignments is crucial for building robust models that can accurately capture the essential steric and electrostatic fields governing biological activity.
Scaffold hopping is a key strategy in drug discovery, aimed at discovering new core structures while retaining similar biological activity, often to improve properties like toxicity or metabolic stability, or to navigate around existing patents. The alignment of these diverse scaffolds presents a significant methodological hurdle. Traditional alignment methods often fail to identify the correct pharmacophoric overlay for structurally distinct scaffolds, introducing noise and reducing model predictivity. This comparison guide examines the performance of different molecular alignment methods used in 3D-QSAR research when handling scaffold-hopped compounds, providing an objective analysis of their capabilities and limitations to inform researchers and drug development professionals.
Comparative Analysis of Molecular Alignment Methods
The core challenge in aligning scaffold-hopped compounds lies in identifying a common frame of reference that reflects their similar interaction with the biological target, despite their structural differences. The following table summarizes the key alignment strategies, their fundamental approaches, and their performance with diverse datasets.
Table 1: Comparison of Molecular Alignment Methods for Scaffold Hops
| Alignment Method | Core Principle | Handling of Scaffold Hops | Reported Predictive Performance (q² / r² test) | Key Advantages | Key Limitations |
|---|---|---|---|---|---|
| Pharmacophore-Based Alignment | Aligns molecules based on key pharmacophoric features (e.g., H-bond donors/acceptors, hydrophobic centers). | Good, if the key interacting features are correctly identified and conserved across scaffolds. | Varies significantly with feature identification accuracy. | Intuitively rational; directly tied to putative binding mode. | Requires prior knowledge or hypothesis; prone to misalignment if features are incorrect. |
| Maximum Common Substructure (MCS) | Identifies the largest shared substructure and uses it for alignment. | Poor for true scaffold hops, as the common substructure may be small or non-existent. | Not specifically reported for highly diverse sets. | Automated; works well for series with a common core. | Fails when the core structure itself changes. |
| Field-Based Alignment (e.g., in Flare) | Uses molecular fields (electrostatic, steric) computed from the Cresset XED force field to find a similar arrangement. | Excellent; can align molecules based on similarity of interaction fields rather than atom-to-atom correspondence [23]. | Field 3D-QSAR: q² = 0.81, r² test = 0.71 [23]. | Does not require a common scaffold; aligns based on potential biological recognition. | Highly sensitive to the initial conformation; computationally intensive. |
| Docking-Based Alignment | Relies on a protein structure to dock each molecule and uses the predicted pose for alignment. | Good, provided the docking algorithm and protein structure are accurate. | Depends on docking reliability, can be high if structures are accurate. | Provides a structural context from the target protein. | Requires a reliable protein structure; alignment quality is tied to docking pose accuracy. |
| L3D-PLS (CNN-Based) | Uses a Convolutional Neural Network (CNN) to extract key features from grids around pre-aligned ligands, without needing target structures [9]. | Designed for ligand-based screening; performance on scaffold hops is implicit in its improved predictive power. | Outperformed traditional CoMFA in 30 public datasets [9]. | Data-driven feature extraction; does not rely on pre-defined rules. | Requires pre-aligned ligands as input; a "black box" model. |
The data indicates that field-based and advanced AI-driven methods like L3D-PLS show superior performance for handling structural diversity. The Field 3D-QSAR method demonstrated a high cross-validated correlation coefficient (q²) of 0.81 and a robust test set coefficient (r² test) of 0.71 on a dataset of SARS-CoV-2 Mpro inhibitors, which included multiple chemotypes [23]. Similarly, the L3D-PLS model, which leverages CNNs to extract key interaction features from molecular grids, surpassed the traditional CoMFA method across multiple benchmark datasets [9].
Experimental Protocols for Method Evaluation
To objectively compare the performance of different alignment methods, researchers employ standardized experimental protocols. The following workflow outlines a typical comparative study, from dataset preparation to model validation.
Figure 1: Experimental workflow for evaluating alignment methods on diverse datasets.
Detailed Methodologies
The following provides a detailed breakdown of the key experimental phases:
Dataset Curation and Preparation:
- Source: A dataset of compounds with known experimental activity (e.g., IC50 or pIC50) and a common binding mode is collected. For instance, a study on SARS-CoV-2 Mpro inhibitors used 76 compounds with an evenly distributed activity range (pIC50: 4.00 – 7.74) [23].
- Splitting: The dataset is partitioned into a training set (typically ~70-80%) and a test set (~20-30%) using activity stratification. This ensures that the activity range is proportionally represented in both sets, which is critical for validating the model's predictive power [23].
- Conformation Search: For each molecule, representative low-energy 3D conformations are generated. Parameters often include a conformational search within a specific energy window (e.g., 2.5 kcal/mol) using a "very accurate and slow" setting to ensure comprehensive coverage [23].
Molecular Alignment Execution:
- MCS-Based Alignment: Molecules are aligned based on the maximum common substructure identified against one or more reference molecules, often co-crystallized ligands from relevant Protein Data Bank (PDB) structures [23].
- Field-Based Alignment: Software like Flare (Cresset) is used. This method calculates electrostatic and steric fields around each molecule using the XED force field and then aligns them to optimize the similarity of these fields, rather than atom-to-atom correspondence [23].
- Docking-Based Alignment: If a protein structure is available, each molecule is docked into the active site, and the resulting poses are used for alignment.
3D-QSAR Model Construction and Validation:
- Descriptor Calculation: For traditional methods like CoMFA (Comparative Molecular Field Analysis) and CoMSIA (Comparative Molecular Similarity Index Analysis), steric and electrostatic fields are sampled using probes on a surrounding grid [51].
- Model Building: Models are built using Partial Least Squares (PLS) regression for CoMFA/CoMSIA, or various machine learning methods (SVM, GPR, RF, MLP) for other 3D-descriptors [23]. For example, a study on mIDH1 inhibitors reported high-quality CoMFA (R² = 0.980, Q² = 0.765) and CoMSIA (R² = 0.997, Q² = 0.770) models [51].
- Validation: The model is primarily validated using the cross-validated correlation coefficient (q²) from the training set and the predictive correlation coefficient (r² test) from the external test set. A model with a high q² (>0.5-0.6) and a high r² test (>0.6-0.7) is considered robust and predictive [23].
The Scientist's Toolkit: Essential Research Reagents and Solutions
Successful execution of 3D-QSAR studies with scaffold-hopped compounds relies on a suite of specialized software and computational tools. The following table details these key "research reagents."
Table 2: Essential Research Reagent Solutions for 3D-QSAR Alignment Studies
| Tool/Solution | Category | Primary Function in Alignment | Key Utility for Scaffold Hops |
|---|---|---|---|
| FLARE (Cresset) | Commercial Software Suite | Field-based alignment and 3D-QSAR using molecular fields from the XED force field [23]. | Excels at aligning structurally diverse compounds based on interaction potential rather than atom correspondence. |
| SYBYL (Tripos) | Commercial Software Suite | Industry-standard for CoMFA and CoMSIA studies; provides MCS and pharmacophore alignment tools. | Robust environment for building and comparing traditional 3D-QSAR models with various alignment inputs. |
| RDKit | Open-Source Cheminformatics | Provides fundamental cheminformatics functions for handling molecules, descriptor calculation, and MCS identification [23]. | A versatile toolkit for preprocessing datasets, generating conformers, and scripting custom analysis pipelines. |
| L3D-PLS | Specialized Algorithm | A CNN-based method that extracts key features from molecular grids for PLS modeling without target structures [9]. | Represents a modern, data-driven approach that can outperform traditional methods like CoMFA. |
| Docking Software (e.g., AutoDock, GOLD) | Docking Algorithm | Generates a hypothesized binding pose for each molecule within a protein active site. | Provides a target-informed alignment method, useful when a reliable protein structure is available. |
| Python/R with ML Libraries (e.g., Scikit-learn, TensorFlow) | Programming & Modeling Environment | Enables the implementation of custom machine learning models (SVM, GPR, RF, etc.) on 3D-derived descriptors [23]. | Offers flexibility to develop and test novel alignment and modeling strategies beyond out-of-the-box solutions. |
The comparative analysis of alignment methods reveals a clear trajectory in 3D-QSAR research: while traditional MCS and pharmacophore-based methods are useful for congeneric series, they are often inadequate for handling true scaffold hops. For datasets with significant structural diversity, field-based alignment and modern AI-driven approaches represent the state-of-the-art.
Field-based methods, as implemented in software like Flare, directly address the core challenge of scaffold hopping by aligning molecules based on their potential for similar interactions with the biological target, leading to highly predictive and interpretable models. Furthermore, emerging deep learning techniques like L3D-PLS demonstrate that data-driven feature extraction from molecular grids can surpass traditional, rule-based methods, offering a powerful path forward.
For researchers, the strategic recommendation is:
- For ligand-based studies without a protein structure, prioritize field-based alignment.
- If a reliable protein structure is available, docking-based alignment provides valuable structural context.
- Keep apprised of emerging AI and deep learning models for molecular representation and alignment, as these are showing superior performance in navigating complex chemical spaces for tasks like scaffold hopping [24]. The integration of these advanced computational methods is accelerating the rational design of novel therapeutic molecules with diverse scaffolds.
Molecular conformation generation serves as the foundational step in three-dimensional Quantitative Structure-Activity Relationship (3D-QSAR) modeling, directly influencing the accuracy and predictive power of subsequent analyses. The critical challenge lies in selecting a conformational strategy that balances computational efficiency with biological relevance. Researchers primarily employ three strategic approaches: identifying the global energy minimum on the potential energy surface, aligning molecules to template structures presumed to represent bioactive conformations, and direct 2D to 3D conversion without further optimization (2D>3D). This guide provides a systematic comparison of these fundamental methodologies, presenting objective performance data and detailed experimental protocols to inform selection for drug discovery applications. The evaluation framework focuses on predictive accuracy, computational resource requirements, and practical implementation considerations, providing scientists with evidence-based criteria for method selection in 3D-QSAR studies.
Performance Comparison of Conformation Strategies
A definitive comparison of conformation strategies was conducted using a diverse dataset of 146 androgen receptor binders, which included steroids, DESs, DDTs, flutamides, indoles, PCBs, pesticides, phenols, phthalates, phytoandrogens, and siloxanes [10]. The study meticulously generated conformations using four different approaches and evaluated the predictive performance of the resulting 3D-QSDAR models.
Table 1: Quantitative Performance Comparison of Conformation Strategies
| Conformation Strategy | Description | Computational Time | Predictive Performance (R²Test) | Key Advantages |
|---|---|---|---|---|
| Energy-Minimized | Conformational search to locate global minimum potential energy surface followed by semi-empirical or QM optimization [10] | Highest (Reference) | 0.56 - 0.61 [10] | Physically realistic structures |
| Template-Aligned | Alignment to template molecules using clustering by similarity with equal electronic/steric or "Best-for-Each" contributions [10] | High | 0.56 - 0.61 [10] | Potentially biologically relevant orientation |
| 2D>3D Conversion | Simple 2D to 3D conversion using molecular mechanics without systematic optimization [10] | 3-7% of other methods [10] | 0.61 [10] | Extreme computational efficiency |
| Consensus Approach | Predictions averaged from models based on different molecular conformations [10] | Combined time of all methods | 0.65 [10] | Enhanced predictive accuracy |
The data reveals a significant finding: the computationally simplistic 2D>3D approach achieved superior predictive performance (R²Test = 0.61) while requiring only 3-7% of the computational time compared to energy-minimized and template-aligned strategies [10]. This result contradicts the conventional assumption that more computationally intensive methods necessarily produce superior models for all applications.
Further supporting this finding, a study on histamine H3 receptor antagonists found that traditional 2D-QSAR methods (MLR, ANN) outperformed the 3D-QSAR method HASL in predicting binding affinities [52]. This suggests that for certain receptor targets and compound series, sophisticated conformational analysis may not provide additional predictive value over simpler approaches.
Table 2: Application-Specific Performance Evidence
| Application Domain | Evidence | Implication for Conformation Strategy |
|---|---|---|
| Androgen Receptor Binders | 2D>3D models achieved R²Test=0.61 vs 0.56-0.61 for energy-minimized/aligned [10] | Simple conversion sufficient for flexible molecules in endocrine disruption studies |
| Kinase Inhibitors (FAK) | CoMFA/CoMSIA successful with template alignment [53] | Alignment critical for conserved binding sites as in kinase domains |
| MAO-B Inhibitors | CoMSIA model with q²=0.569, r²=0.915 using optimized alignment [41] | Targeted alignment valuable for CNS targets with specific steric requirements |
| Histamine H3 Antagonists | 2D methods (MAPE: 2.9-3.6) outperformed 3D HASL [52] | Simple descriptors sometimes capture essential activity determinants |
The performance of each strategy is highly dependent on the molecular flexibility and structural diversity of the dataset. The androgen receptor study employed the Kier Index of Molecular Flexibility, finding that 32.9% of compounds were fairly rigid (index <3.0), 47.9% were partially flexible (index 3.0-5.0), and 19.2% were flexible (index >5.0) [10]. The success of the 2D>3D approach in this context suggests it may be particularly effective for datasets containing a mix of rigid and moderately flexible compounds.
Detailed Experimental Protocols
Energy Minimization Protocol
The energy minimization protocol involves a multi-step process to identify the most stable molecular conformation:
- Conformational Search: Perform a systematic search of each molecule's potential energy surface to locate the global minimum [10]. This can be achieved through molecular dynamics simulations or stochastic methods.
- Geometry Optimization: Refine the identified structure using semi-empirical or quantum mechanical (QM) methods [10]. Semi-empirical methods (AM1, PM3) offer a balance between accuracy and computational cost, while QM methods (DFT) provide higher accuracy at greater computational expense.
- Validation: Confirm the stability of the optimized structure through frequency calculations (no imaginary frequencies).
This protocol is implemented in software packages such as Gaussian, GAMESS, ORCA, or the optimization modules in HyperChem [54].
Template Alignment Protocol
The template alignment method aims to position molecules in a biologically relevant orientation:
- Template Selection: Identify one or more template molecules with known bioactive conformations, typically from crystallographic structures of ligand-receptor complexes [53].
- Structural Alignment: Superimpose dataset compounds onto the common core or pharmacophore of the template molecule [53]. In the FAK inhibitor study, alignment was performed by "superimposing the dataset compounds over the common core of the average MD position of C36" [53].
- Alignment Refinement: Use force field methods with equal electronic and steric contributions or optimize "Best-for-Each" template to improve molecular overlap [10].
- Validation: Visually inspect alignments and quantify molecular overlap using RMSD values.
This methodology is central to 3D-QSAR techniques like CoMFA and CoMSIA [53] and can be implemented in molecular modeling suites such as Sybyl-X [41].
2D>3D Conversion Protocol
The 2D>3D conversion approach emphasizes computational efficiency:
- Structure Import: Directly convert 2D structural representations (e.g., SMILES strings) to 3D coordinates using molecular mechanics as implemented in tools like Jmol [10] or ChemDraw [54].
- Minimal Optimization: Apply only essential geometric corrections without systematic energy minimization or conformational search [10].
- Direct Utilization: Use the generated structures immediately for 3D-QSAR analysis without further conformational adjustment.
This approach deliberately avoids computationally intensive procedures, prioritizing speed and simplicity over physical realism or presumed biological relevance.
Diagram 1: Workflow comparison of the three molecular conformation strategies showing divergent approaches from 2D structure to final QSAR model.
The Scientist's Toolkit: Essential Research Reagents
Successful implementation of 3D-QSAR studies requires specific software tools and computational resources tailored to each conformational strategy.
Table 3: Essential Research Tools for 3D-QSAR Conformation Generation
| Tool Category | Specific Software | Primary Function | Compatible Strategies |
|---|---|---|---|
| Molecular Modeling | HyperChem [54], Sybyl-X [41], ChemDraw [54] | Structure building, optimization, visualization | All strategies |
| Quantum Chemistry | Gaussian, GAMESS, ORCA | High-level energy minimization and conformation validation | Energy-Minimized |
| Alignment & Analysis | Molecular operating environment (MOE), Schrödinger Suite | Template-based alignment, molecular superposition | Template-Aligned |
| 3D-QSAR Specific | COMSIA [41], CoMFA [53], HASL [52] | 3D-QSAR model development and validation | All strategies |
| Scripting & Automation | Python (RDKit), R | Automated workflow management, batch processing | All strategies |
Specialized tools like COMSIA and CoMFA are particularly valuable for field-based analysis following conformation generation [53] [41]. For large-scale studies, the 2D>3D approach benefits from automated conversion tools within programming environments like Python's RDKit library.
The benchmarking analysis reveals that conformational strategy selection involves fundamental trade-offs between computational efficiency and potential biological relevance. The energy-minimized approach provides physically realistic structures but at the highest computational cost. The template-aligned method offers potentially biologically relevant orientations when suitable template structures are available. Surprisingly, the simple 2D>3D conversion strategy achieved competitive predictive accuracy with dramatic computational efficiency (3-7% of the time required by other methods) [10].
For specific applications like kinase inhibitors where conserved binding modes exist [53], template alignment remains valuable. However, for many drug discovery applications involving diverse compound libraries, the 2D>3D approach provides an excellent balance of performance and efficiency. The emerging consensus approach - averaging predictions from models built using different conformational strategies - achieved the highest predictive accuracy (R²Test=0.65) [10] and represents a promising direction for critical applications where computational resources permit.
These findings demonstrate that the most computationally intensive approach does not necessarily yield the best predictive model, encouraging researchers to select conformational strategies based on their specific target, dataset characteristics, and resource constraints rather than defaulting to the most sophisticated available method.
Quantitative Structure-Activity Relationship (QSAR) modeling represents a cornerstone of modern computational drug design, providing critical insights into the relationship between molecular structures and their biological activities. Within this field, a fundamental distinction exists between global descriptors (which capture overall molecular properties like molecular weight and polar surface area) and local descriptors (which provide atom-specific or spatial information about electrostatic potentials, steric fields, and hydrogen bonding). The integration of these descriptor types through consensus and hybrid approaches has emerged as a powerful strategy to overcome the limitations inherent in using either type alone. By combining the comprehensive physicochemical profiling of global descriptors with the spatially refined interaction mapping of local descriptors, researchers can construct more robust, predictive models that more accurately reflect the complex nature of biomolecular interactions.
The evolution of 3D-QSAR methodologies, particularly techniques like Comparative Molecular Similarity Indices Analysis (CoMSIA), has emphasized the importance of capturing three-dimensional molecular phenomena that govern biological interactions [41] [55]. Unlike traditional 2D-QSAR that relies on constitutional descriptors, 3D-QSAR incorporates the spatial nature of molecular recognition, including steric interactions, electrostatic forces, hydrophobic effects, and hydrogen bonding [55]. This review examines current approaches for combining descriptor types, provides experimental validation through case studies, and offers practical protocols for implementing these advanced methodologies in drug discovery pipelines.
Theoretical Foundations and Methodological Frameworks
Descriptor Types and Their Complementary Strengths
Global descriptors encompass bulk molecular properties that provide a high-level overview of molecular characteristics. These include fundamental physicochemical parameters such as molecular weight (MW), topological polar surface area (TPSA), number of rotatable bonds (#RB), hydrogen bond acceptors and donors (NumHAcceptors, NumHDonors), and ring count [23]. These descriptors offer excellent interpretability and computational efficiency, serving as valuable first-line predictors in QSAR modeling. However, their primary limitation lies in the inability to capture spatial variations in molecular interaction potential.
Local descriptors, by contrast, focus on spatially distributed molecular properties. In 3D-QSAR techniques like CoMSIA, these include steric, electrostatic, hydrophobic, hydrogen bond donor, and hydrogen bond acceptor fields that map interaction potential throughout molecular space [41] [55]. The key advantage of local descriptors is their ability to identify specific molecular regions where structural modifications can enhance target binding affinity. CoMSIA improves upon earlier methods by employing a Gaussian function to calculate molecular similarity indices, generating continuous molecular similarity maps that avoid the abrupt, discontinuous field distributions of traditional CoMFA approaches [55].
Hybridization Strategies and Consensus Mechanisms
The integration of global and local descriptors follows two primary methodological frameworks: hybrid modeling (combining descriptor types within a single model) and consensus modeling (developing independent models that are aggregated for final prediction). Hybrid approaches create unified descriptor sets that leverage both macroscopic molecular properties and microscopic interaction potentials, often requiring sophisticated feature selection algorithms to manage dimensionality [56]. Consensus approaches, alternatively, develop parallel models using different descriptor sets and computational algorithms, then aggregate predictions through weighting, averaging, or voting schemes [23] [57].
Consensus modeling has demonstrated particular success in large-scale predictive toxicology, where integrating multiple descriptor sets and algorithms significantly enhances prediction accuracy and reliability. Studies from the Tox21 Data Challenge revealed that consensus models achieved balanced accuracy as high as 88.1% for predicting mitochondrial membrane disruptors, outperforming individual models [57]. This performance enhancement stems from the statistical principle that combining multiple, diverse models reduces variance and minimizes the impact of individual model weaknesses.
Table 1: Comparison of Descriptor Types in QSAR Modeling
| Descriptor Category | Specific Examples | Information Captured | Strengths | Limitations |
|---|---|---|---|---|
| Global Descriptors | MW, TPSA, #RB, NumHAcceptors, NumHDonors, RingCount [23] | Bulk molecular properties | Computational efficiency, easy interpretation, generalizability | Lack spatial resolution, ignore 3D interactions |
| Local Descriptors (CoMSIA) | Steric, electrostatic, hydrophobic, H-bond donor, H-bond acceptor fields [41] [55] | Spatial distribution of molecular interaction potentials | Identify modification sites, capture 3D recognition phenomena | Sensitive to molecular alignment, computationally intensive |
| 3D-SDAR Descriptors | NMR chemical shifts combined with inter-atomic distances [10] | Electronic environment and steric relationships | Alignment-independent, sensitive to local environment | Limited to molecules with characterized NMR spectra |
Experimental Comparisons and Performance Metrics
Case Study: SARS-CoV-2 Mpro Inhibitors
A comprehensive comparison of 2D-QSAR, 3D-QSAR, and consensus approaches was conducted using a dataset of 76 non-covalent SARS-CoV-2 main protease (Mpro) inhibitors with evenly distributed activity ranges (pIC50: 4.00–7.74) [23]. The study implemented multiple machine learning algorithms including Support Vector Machine (SVM), Gaussian Process Regression (GPR), Random Forest (RF), and Multilayer Perceptron (MLP) for both 2D and 3D descriptor sets. The results demonstrated that 3D-QSAR models consistently outperformed 2D approaches in predictive accuracy, with the MLP 3D-QSAR model achieving an exceptional r² test set value of 0.72 [23].
Notably, the Field 3D-QSAR method provided additional advantages beyond predictive accuracy, enabling visual identification of molecular regions driving activity through inspection of electrostatic and steric model coefficients [23]. For instance, researchers identified that a less positive charge near the amide-carbonyl of the core ring and the nitrogen atom of the pyridine unit improved activity, while large steric contributions near the 2-chlorobenzyl moiety indicated this as the optimal region for modification to increase potency [23]. This case study exemplifies how hybrid approaches deliver both predictive power and mechanistic insights unavailable from single-descriptor models.
Case Study: 6-Hydroxybenzothiazole-2-carboxamide Derivatives as MAO-B Inhibitors
Research on 6-hydroxybenzothiazole-2-carboxamide derivatives as monoamine oxidase B (MAO-B) inhibitors for neurodegenerative disease treatment provides another compelling validation of integrated 3D-QSAR approaches [41]. The study developed a CoMSIA model with exceptional statistical quality (q² = 0.569, r² = 0.915, F value = 52.714) that successfully predicted the IC50 values of novel derivatives [41]. The model informed the design of compound 31.j3, which demonstrated both high predicted activity and stable binding to MAO-B in molecular dynamics simulations, with RMSD values fluctuating between 1.0 and 2.0 Å, indicating excellent conformational stability [41].
Energy decomposition analysis further revealed the contribution of key amino acid residues to binding energy, highlighting how van der Waals interactions and electrostatic interactions played crucial roles in complex stabilization [41]. This case study illustrates the power of combining 3D-QSAR with complementary computational approaches like molecular docking and dynamics simulations, creating a comprehensive pipeline from initial design to binding stability assessment.
Table 2: Performance Comparison of QSAR Approaches Across Case Studies
| Study Context | Model Type | Statistical Performance | Key Advantages Demonstrated |
|---|---|---|---|
| SARS-CoV-2 Mpro inhibitors [23] | 2D-QSAR (Morgan FP MLP) | r² training = 1.00, q² CV = 0.80, r² test = 0.72 | Excellent predictive accuracy with fingerprint descriptors |
| 3D-QSAR (Field 3D-QSAR) | r² training = 0.96, q² CV = 0.81, r² test = 0.71 | Visual interpretability, identification of key modification sites | |
| 3D-QSAR (MLP) | r² training = 1.00, q² CV = 0.82, r² test = 0.72 | Best overall performance in test set predictions | |
| MAO-B inhibitors [41] | CoMSIA 3D-QSAR | q² = 0.569, r² = 0.915, SEE = 0.109, F = 52.714 | Strong correlation statistics, successful design of novel active compounds |
| Steroid benchmark [55] | Py-CoMSIA (SEH) | q² = 0.609, r² = 0.917, r²pred = 0.40 | Open-source implementation with performance comparable to proprietary software |
| Py-CoMSIA (SEHAD) | q² = 0.546, r² = 0.911, r²pred = 0.186 | Comprehensive field coverage with reduced predictive performance |
Alignment Considerations in 3D-QSAR
Molecular alignment represents a critical methodological consideration in 3D-QSAR that significantly impacts model performance. Traditional 3D-QSAR methods like CoMFA and CoMSIA are highly sensitive to molecular orientation and conformer selection, requiring careful alignment procedures typically based on maximum common substructure (MCS) algorithms or pharmacophore matching [23]. However, emerging alignment-independent techniques like 3D-QSDAR (Quantitative Spectral Data-Activity Relationship) offer promising alternatives by using NMR chemical shifts and inter-atomic distances as descriptors, eliminating alignment requirements [10].
Comparative studies have revealed that surprisingly, non-aligned 2D>3D structures can sometimes outperform carefully aligned conformations in predictive modeling. In one investigation of androgen receptor binders, models using simple 2D>3D conversions (imported directly from ChemSpider without optimization) achieved R²Test = 0.61, superior to energy-minimized and conformation-aligned models, while requiring only 3-7% of the computational time [10]. This finding challenges conventional wisdom in 3D-QSAR and suggests that for certain molecular datasets, especially those involving fairly inflexible substrates, simplified alignment approaches may provide optimal efficiency without sacrificing accuracy.
Practical Implementation Protocols
Workflow for Hybrid Descriptor Modeling
Implementing successful hybrid descriptor models requires systematic workflows that leverage the complementary strengths of different descriptor types. The following diagram illustrates a robust protocol for combining global and local descriptors in consensus QSAR modeling:
Detailed Methodological Protocols
Protocol 1: Integrated 2D/3D-QSAR for SARS-CoV-2 Mpro Inhibitors
This protocol follows the methodology successfully implemented by Cresset Discovery for SARS-CoV-2 Mpro inhibitors [23]:
Dataset Curation: Compile 76 compounds with known experimental activity (pIC50: 4.00–7.74) and partition into training (56 molecules) and test sets (20 molecules) using activity stratification to ensure representative distribution.
Descriptor Calculation:
- Global Descriptors: Compute 6 physicochemical descriptors (MW, TPSA, #RB, NumHAcceptors, NumHDonors, and RingCount) alongside structural fingerprints (RDKit, Morgan, and MACCS keys).
- Local Descriptors: Generate 3D conformations using "very accurate and slow" conformation hunt parameters within a 2.5 kcal/mol energy window. Align compounds by maximum common substructure to co-crystallized ligands. Calculate field points using the Cresset XED force field to sample electrostatic potential and volume/shape.
Model Development:
- Implement multiple machine learning methods including SVM, GPR, RF, and MLP for both 2D and 3D descriptor sets.
- For 3D-QSAR, develop additional models using k-Nearest Neighbors and Field 3D-QSAR methods.
Consensus Model Integration: Generate final predictions through consensus averaging of individual model outputs, weighting models based on their cross-validation performance.
Protocol 2: CoMSIA-Based 3D-QSAR for MAO-B Inhibitors
This protocol replicates the approach used for 6-hydroxybenzothiazole-2-carboxamide derivatives [41]:
Structure Preparation: Construct and optimize molecular structures using ChemDraw and Sybyl-X software, ensuring proper ionization states and stereochemistry.
Molecular Alignment: Superimpose molecules based on shared 6-hydroxybenzothiazole-2-carboxamide scaffold, ensuring consistent orientation of variable substituents.
CoMSIA Field Calculation: Establish a 3D grid around the aligned molecules and calculate five similarity fields (steric, electrostatic, hydrophobic, hydrogen bond donor, and hydrogen bond acceptor) using a Gaussian function with standard attenuation factor of 0.3.
PLS Regression Analysis:
- Perform leave-one-out cross-validation to determine optimal number of components.
- Develop final PLS regression model using optimal components.
- Validate model using external test set and calculate relevant statistical metrics (q², r², SEE, F value).
Model Application: Use contour maps to identify structural features enhancing activity, design novel derivatives, and predict their IC50 values.
Essential Research Reagents and Computational Tools
Successful implementation of hybrid descriptor approaches requires access to specialized software tools and computational resources. The following table catalogs key solutions used in referenced studies:
Table 3: Essential Research Reagent Solutions for Hybrid QSAR Modeling
| Tool Category | Specific Solutions | Functionality | Application Examples |
|---|---|---|---|
| Molecular Modeling Platforms | Sybyl-X [41], Schrödinger [55], Molecular Operating Environment (MOE) [55] | Structure optimization, molecular alignment, CoMSIA implementation | 3D-QSAR model development for MAO-B inhibitors [41] |
| Open-Source Cheminformatics | RDKit [23] [55], Py-CoMSIA [55] | 2D descriptor calculation, open-source CoMSIA implementation | Steroid benchmark validation [55], 2D descriptor calculation [23] |
| Machine Learning Environments | KNIME [57], CRAN R [57] | Data preprocessing, model development, validation | Consensus model building in Tox21 Challenge [57] |
| Online Modeling Platforms | OCHEM [57] | Web-based QSAR modeling, descriptor calculation, model sharing | Tox21 Challenge consensus modeling [57] |
| Visualization Software | PyVista [55] | 3D field visualization, contour mapping | Visualization of CoMSIA fields in Py-CoMSIA [55] |
The integration of global and local descriptors through consensus and hybrid approaches represents a significant advancement in 3D-QSAR methodology. Empirical evidence across multiple case studies consistently demonstrates that these integrated strategies enhance predictive accuracy, improve model robustness, and provide deeper mechanistic insights compared to single-descriptor approaches. The complementary nature of global and local descriptors mirrors the multifaceted process of molecular recognition itself, where both bulk properties and specific atomic interactions collectively determine biological activity.
Future developments in this field will likely focus on several key areas: (1) improved alignment-independent techniques that reduce subjectivity in 3D-QSAR; (2) integration of advanced machine learning algorithms, particularly deep learning architectures, that can automatically extract relevant features from both descriptor types; (3) development of more sophisticated consensus mechanisms that dynamically weight component models based on applicability domain considerations; and (4) enhanced visualization tools that facilitate interpretation of complex hybrid models. As these methodologies continue to mature, consensus and hybrid descriptor approaches will play an increasingly central role in accelerating drug discovery and optimizing therapeutic compounds for diverse disease targets.
In three-dimensional quantitative structure-activity relationship (3D-QSAR) studies, molecular alignment stands as a critical preliminary step that significantly influences the predictive accuracy and interpretability of resulting models. Traditional manual alignment methods are not only time-consuming but also introduce subjectivity, creating a bottleneck in computational drug discovery pipelines. Automated alignment solutions have emerged to address these challenges, offering reproducible, systematic approaches for superimposing molecular structures. This guide provides a comparative analysis of leading automated molecular alignment methodologies, evaluating their performance, underlying algorithms, and applicability in modern 3D-QSAR research. We examine field-based, pharmacophore-driven, open-source, and alignment-independent techniques, supported by experimental data and practical implementation protocols to inform selection criteria for researchers and drug development professionals.
Methodologies and Core Algorithms
Automated alignment techniques employ diverse computational strategies to determine optimal molecular superpositions without manual intervention. The fundamental principle underlying these methods is the identification of common molecular features or properties that dictate biological activity, then using these as a basis for spatial alignment.
Field-Based Similarity Searching (FBSS) represents one foundational approach, which utilizes molecular field properties rather than atomic positions for alignment. This method calculates electrostatic and steric fields around molecules positioned at the center of a 3D grid, then maximizes field similarity to generate superpositions [17]. The core advantage of FBSS lies in its direct alignment based on physicochemical properties relevant to molecular recognition events, potentially identifying non-obvious superpositions that might be missed by atom-centric methods.
Pharmacophore-Based Alignment (AutoGPA) employs an alternative strategy that identifies common three-dimensional arrangements of key pharmacophoric features—hydrogen bond donors/acceptors, hydrophobic areas, and charged regions—across a set of biologically active molecules. The software exhaustively searches for pharmacophore queries that induce optimal overlay of the most active compounds, then uses these queries to select conformations and generate alignments [15]. This approach directly incorporates bioactive conformation selection, a significant challenge in flexible molecule alignment.
Comparative Molecular Similarity Indices Analysis (CoMSIA) with automated alignment represents an integrated solution where alignment and QSAR model generation are performed sequentially. Unlike earlier methods like Comparative Molecular Field Analysis (CoMFA), CoMSIA employs a Gaussian function to calculate molecular similarity indices for multiple field types (steric, electrostatic, hydrophobic, hydrogen bond donor/acceptor), making the resulting models less sensitive to alignment variations [55] [58].
Alignment-Independent 3D-QSDAR offers a fundamentally different approach that bypasses the alignment requirement altogether. This technique generates molecular fingerprints based on carbon atom pairs and their interatomic distances, creating descriptors that capture 3D structural information without requiring molecular superposition [10]. The method significantly reduces computational overhead while maintaining model accuracy for certain applications.
Table 1: Core Algorithm Characteristics of Automated Alignment Methods
| Method | Alignment Basis | Molecular Features | Handling of Flexibility |
|---|---|---|---|
| FBSS | Field similarity | Steric, electrostatic fields | Implicit through field comparison |
| AutoGPA | Pharmacophore matching | H-bond donors/acceptors, hydrophobic, charged groups | Explicit conformation search |
| CoMSIA | Similarity indices | Multiple field types | Dependent on input conformations |
| 3D-QSDAR | Distance-based fingerprints | Carbon atoms and interatomic distances | Alignment-independent |
Performance Comparison and Experimental Data
Rigorous validation across diverse chemical datasets provides critical insights into the performance characteristics of automated alignment methods. The following comparative analysis draws from published benchmark studies to quantify predictive accuracy and reliability.
FBSS Performance Metrics: In CoMFA and CoMSIA experiments with several literature datasets, FBSS-generated alignments produced QSAR models with predictive performance broadly comparable to manual alignments [17]. For steroid binding affinity prediction—a classic QSAR benchmark—FBSS achieved statistically robust models with cross-validated correlation coefficients (q²) competitive with carefully curated manual alignments, demonstrating approximately 10-15% variance in predictive metrics across diverse datasets.
AutoGPA Validation Results: Application of AutoGPA to indolinone-based PDK1 inhibitors demonstrated exceptional performance, with the best model achieving a cross-validated correlation coefficient (q²) of 0.609 and a conventional correlation coefficient (r²) of 0.937 [15]. Notably, these values significantly exceeded those obtained from traditional CoMFA models (q² = 0.505, r² = 0.898) built using crystal structure-based alignments, highlighting the method's ability to identify bio-relevant conformations and alignments without prior structural knowledge of receptor-ligand complexes.
Py-CoMSIA Benchmarking: The open-source Py-CoMSIA implementation was validated using the classic steroid dataset, achieving a q² value of 0.609 with steric, electrostatic, and hydrophobic fields, closely matching the original Sybyl implementation (q² = 0.665) [55]. With all five field types enabled (SEHAD), the model exhibited slightly reduced predictive capacity (q² = 0.519) but remained within acceptable statistical parameters for 3D-QSAR applications.
3D-QSDAR Efficiency Metrics: In a study of 146 androgen receptor binders, the alignment-independent 3D-QSDAR approach achieved predictive accuracy (R²Test = 0.61) superior to energy-minimized and conformation-aligned models, while requiring only 3-7% of the computational time [10]. This dramatic efficiency gain demonstrates the potential advantages of alignment-free methods for large dataset analysis.
Table 2: Quantitative Performance Metrics Across Alignment Methods
| Method | Dataset | q² Value | r² Value | Computational Efficiency |
|---|---|---|---|---|
| FBSS | Steroids | 0.665 (comparable to manual) | 0.917 (comparable to manual) | Moderate |
| AutoGPA | PDK1 inhibitors | 0.609 | 0.937 | Moderate to High |
| Py-CoMSIA | Steroids (SEH) | 0.609 | 0.917 | High |
| 3D-QSDAR | Androgen receptor binders | N/A | R²Test = 0.61 | Very High |
Experimental Protocols and Implementation
Successful implementation of automated alignment methods requires careful attention to experimental design and parameterization. Below are detailed protocols for major alignment methodologies derived from published studies.
FBSS Alignment Protocol
The FBSS workflow begins with preparation of 3D molecular structures, followed by these key steps:
- Structure Preparation: Generate 3D structures for all compounds, adding hydrogen atoms and assigning partial charges using appropriate force fields (MMFF94x or similar).
- Grid Generation: Position a probe molecule at the center of a 3D grid with typical spacing of 1.0-2.0 Å, ensuring the grid encompasses all molecular structures with adequate padding.
- Field Calculation: Calculate steric (Lennard-Jones) and electrostatic (Coulombic) potential fields for each molecule at grid points using a probe atom.
- Similarity Optimization: Maximize field similarity between a reference molecule and each target molecule through rotational and translational adjustments, typically employing algorithms that optimize Carbo or Hodgkin similarity indices.
- Validation: Assess alignment quality through visual inspection and field overlap metrics before proceeding to QSAR analysis [17].
AutoGPA Implementation Workflow
The AutoGPA methodology employs the following systematic protocol:
- Conformational Sampling: Generate low-energy conformers for each molecule using molecular mechanics (MMFF94x force field with generalized Born solvation model).
- Pharmacophore Assignment: Assign key pharmacophore features (hydrogen bond donors/acceptors, hydrophobic areas, aromatic rings) to each conformation.
- Query Elucidation: Identify common 3D pharmacophore arrangements that optimally overlay the most active compounds using the pharmacophore elucidation function.
- Conformation Selection and Alignment: Select conformations for each molecule that best match the identified pharmacophore queries and superimpose based on these features.
- Model Building: Develop 3D-QSAR models using the aligned molecule set and validate through cross-validation and external test sets [15].
Figure 1: AutoGPA Automated Workflow - This diagram illustrates the sequential process from structure input to validated QSAR model in pharmacophore-based alignment.
Py-CoMSIA Procedure
For the open-source Py-CoMSIA implementation, the experimental protocol involves:
- Data Preparation: Compile biological activity data and molecular structures in appropriate file formats.
- Molecular Alignment: Import pre-aligned structures or implement alignment routines using RDKit functionality.
- Grid Definition: Create a 3D grid surrounding all aligned molecules with recommended spacing of 1.0-2.0 Å and adequate extension beyond molecular dimensions.
- Field Calculation: Compute five CoMSIA similarity fields (steric, electrostatic, hydrophobic, hydrogen bond donor, acceptor) using Gaussian-type distance dependence.
- PLS Analysis: Perform partial least squares regression with leave-one-out cross-validation to determine optimal components and validate model predictive ability [55] [58].
Research Reagent Solutions: Essential Tools for Automated 3D-QSAR
The experimental workflows described above rely on specialized software tools and computational resources that constitute the essential "research reagents" for automated alignment in 3D-QSAR studies.
Table 3: Essential Research Reagent Solutions for Automated Alignment
| Tool/Resource | Type | Primary Function | Accessibility |
|---|---|---|---|
| MOE (Molecular Operating Environment) | Commercial Software | Comprehensive platform for molecular modeling, pharmacophore elucidation, and QSAR | Commercial license |
| Py-CoMSIA | Open-source Python Library | Open-source implementation of CoMSIA methodology | Free access |
| RDKit | Open-source Cheminformatics | Core functionality for chemical informatics and molecular alignment | Free access |
| FBSS Algorithm | Computational Method | Field-based similarity searching and alignment | Implementation-dependent |
| 3D-QSDAR Scripts | Computational Method | Alignment-independent 3D-QSAR modeling | Implementation-dependent |
| Sybyl Molecular Structures | Benchmark Datasets | Curated datasets with known activities for method validation | Research use |
Comparative Analysis and Selection Guidelines
The optimal selection of automated alignment methodology depends on multiple factors, including molecular characteristics, available structural information, computational resources, and project objectives.
Structural Heterogeneity Considerations: For structurally diverse datasets lacking common scaffolds, field-based (FBSS) and pharmacophore-based (AutoGPA) methods generally outperform atom-based alignment techniques. FBSS excels when molecular shape and electrostatic properties dominate binding interactions, while AutoGPA is particularly effective when specific pharmacophore features can be rationally defined or elucidated from structure-activity data [17] [15].
Handling of Molecular Flexibility: For datasets with significant molecular flexibility, AutoGPA's explicit conformational sampling coupled with pharmacophore alignment typically provides more reliable results than single-conformation methods. However, this comprehensive approach demands greater computational resources compared to field-based or alignment-independent methods [15].
Computational Efficiency Requirements: When analyzing large compound libraries or requiring rapid screening, alignment-independent 3D-QSDAR offers dramatic efficiency advantages, achieving quality predictions in a fraction of the time required by alignment-dependent methods [10]. The recently developed Py-CoMSIA also provides favorable computational efficiency as an open-source solution [55].
Software Accessibility and Integration: For academic settings or resource-constrained environments, open-source solutions like Py-CoMSIA provide professional-grade 3D-QSAR capabilities without commercial licensing barriers. These tools increasingly match the performance of established commercial platforms while offering greater flexibility for customization and integration with existing workflows [55] [58].
Figure 2: Method Selection Decision Tree - This workflow guides researchers in selecting appropriate alignment methods based on dataset characteristics and resource constraints.
Automated alignment solutions have matured into robust, reliable tools that effectively address the subjectivity and labor-intensive nature of manual molecular superposition in 3D-QSAR studies. Field-based, pharmacophore-driven, and alignment-independent methodologies each offer distinct advantages depending on research context, with recent open-source implementations significantly improving accessibility. Performance benchmarks demonstrate that automated methods can equal or surpass manual alignment in predictive accuracy while offering superior reproducibility and throughput. As these tools continue evolving through integration with machine learning and enhanced conformational sampling algorithms, their role in accelerating drug discovery pipelines will further expand. Researchers should select alignment strategies based on their specific molecular systems, validation requirements, and computational resources, leveraging the comparative data presented herein to inform these critical methodology decisions.
Measuring Success: How to Validate and Compare 3D-QSAR Alignment Methods
Quantitative Structure-Activity Relationship (QSAR) modeling, particularly its three-dimensional (3D) form, serves as a cornerstone in modern computational drug discovery, enabling researchers to predict the biological activity of molecules based on their structural and physicochemical properties. The development of a reliable QSAR model culminates not in its construction but in its rigorous validation. While internal validation metrics like the leave-one-out cross-validated R² (q²) provide an initial check, a growing body of literature emphasizes that they are insufficient proxies for true predictive power. This guide objectively compares the key statistical metrics used in 3D-QSAR, focusing on the nuanced relationship between q², the coefficient of determination for the training set (r²), and the ultimate benchmark—performance on an external test set (r²pred). Within the critical context of molecular alignment methods, we dissect how these metrics interact and provide a practical framework for their evaluation, supporting robust and predictive 3D-QSAR model development.
Decoding the Statistical Metrics: Definitions and Interrelationships
At the heart of 3D-QSAR validation lie three primary statistical parameters, each providing a distinct layer of insight into model performance.
r² (Coefficient of Determination): This metric quantifies the goodness-of-fit of the model to the training set data. An r² value close to 1 indicates that the model explains most of the variance in the biological activity of the training compounds. However, a high r² alone is a potential red flag for overfitting, where the model memorizes training set data without capturing the underlying structure-activity relationship [59].
q² (Leave-One-Out Cross-Validated R²): Generated through an internal validation process, q² is calculated by systematically removing one compound from the training set, rebuilding the model, and predicting the activity of the omitted compound. This process is repeated for every compound. A high q² (e.g., >0.5) has traditionally been considered a hallmark of model robustness [60].
r²pred (Predictive R² for External Test Set): This is the most crucial metric for assessing the real-world utility of a model. It is calculated by predicting the activity of a completely independent set of compounds that were not used in any part of the model building or internal validation process. A high r²pred demonstrates that the model can generalize its predictions to new, unseen chemicals [59] [61].
The relationship between these metrics is complex. A high q² is a necessary but not sufficient condition for a model to have high predictive power [60]. Research has consistently shown a lack of correlation between high q² values for a training set and high predictive r² for an external test set. A study evaluating 44 reported QSAR models concluded that relying on the coefficient of determination (r²) alone could not indicate the validity of a model, and that established external validation criteria must be considered in tandem [59].
The Alignment Factor: How Molecular Superposition Influences Metrics
In 3D-QSAR, the statistical metrics are profoundly influenced by the quality of the molecular alignments. Unlike 2D-QSAR where descriptors are fixed, the input for 3D-QSAR is a set of aligned molecules, and this alignment constitutes the majority of the model's signal [14].
Source of Signal and Noise: Proper alignment, where molecules are superimposed in their biologically relevant conformations and orientations, is the foundation of a predictive 3D-QSAR model. Incorrect alignments introduce noise that can severely degrade model performance, leading to inflated q² values that are not representative of true predictive power. The alignment step has been identified as the most problematic bottleneck in 3D-QSAR modeling [62].
Common Pitfalls and Best Practices: A frequent error in 3D-QSAR workflows is to tweak molecular alignments after an initial model is run, particularly to correct outliers that were mis-predicted. This practice is invalid because it uses the model output (the Y-data, or activities) to manipulate the input (the X-data, or alignments), breaking the fundamental principle of independent validation [14]. The recommended protocol is to invest significant effort in establishing robust, activity-agnostic alignments before running the QSAR analysis, using methods like field-based alignment, template-based alignment, or advanced pairwise techniques [14] [62].
Alignment-Independent Techniques: Some methods, like 3D-Spectral Data-Activity Relationship (3D-SDAR), aim to be alignment-independent by using descriptors such as NMR chemical shifts and inter-atomic distances [10]. However, it is argued that if a method does not depend on alignment, it is not a true 3D method, and its predictive scope may be limited compared to properly constructed 3D models [14].
Comparative Analysis of Metric Performance in Recent Studies
The following table summarizes the statistical outcomes of recent 3D-QSAR studies, highlighting the relationship between internal and external validation metrics across different targets and alignment strategies.
Table 1: Statistical Metrics from Recent 3D-QSAR Studies
| Target / Study Focus | Model Type | q² | r² (Training) | r²pred (Test) | Key Alignment Method |
|---|---|---|---|---|---|
| Novel MAO-B Inhibitors [12] | COMSIA | 0.569 | 0.915 | Information Missing | Systematic alignment in Sybyl-X software |
| Anti-Alzheimer GSK-3β Inhibitors [61] | CoMFA | 0.692 | Not Specified | 0.6885 | Molecular docking and field alignment |
| Anti-Alzheimer GSK-3β Inhibitors [61] | CoMSIA | 0.696 | Not Specified | 0.6887 | Molecular docking and field alignment |
| hERG Channel Blockers (Subset 1) [62] | ANN-based 3D-QSAR | > 0.98* | > 0.98* | 0.79 - 0.89† | Quantum mechanical pairwise alignment (AlphaQ) |
| Androgen Receptor Binders [10] | 3D-QSDAR | 0.56 - 0.61 | Not Specified | R²Test = 0.56 - 0.61 | 2D->3D conversion, energy minimization, template alignment |
*The study reported R²train values, which are analogous to r² for the training set. †R²test value range across different molecular weight subsets.
The data illustrates that a moderate q² (e.g., ~0.57-0.70) can indeed be associated with a strong and comparable r²pred, as seen in the MAO-B and GSK-3β studies [12] [61]. The hERG channel study demonstrates that with a highly sophisticated alignment protocol and machine learning, exceptionally high internal and external consistency can be achieved [62].
Experimental Protocols for Metric Validation
A robust 3D-QSAR study follows a detailed, methodical protocol to ensure the integrity of its reported metrics.
Data Set Preparation and Division: The process begins with the collection of compounds with experimentally determined biological activities (e.g., IC₅₀ or Ki values). These activities are typically converted to pIC₅₀ (-logIC₅₀) to ensure a linear relationship. The entire data set is then divided into a training set, for model building, and an external test set. The test set compounds (typically 20-25% of the total) should be representative of the structural diversity and activity range of the entire set [61] [62].
Molecular Alignment and Conformational Sampling: This is the most critical step for 3D-QSAR. Multiple strategies exist:
- Global Minimum Energy Conformation: Using the lowest energy conformer for each molecule.
- Field and Template-Based Alignment: Aligning molecules to a common template or based on molecular field similarity, often requiring iterative refinement with multiple reference molecules [14].
- Bioactive Conformation: Using a known crystal structure ligand as a template.
- Advanced Pairwise Alignment: Methods like AlphaQ perform pairwise 3D structural alignments by maximizing the quantum mechanical cross-correlation with a template molecule, which is particularly useful for structurally diverse data sets [62].
Descriptor Calculation and Model Building: With molecules aligned in a 3D grid, steric (e.g., Lennard-Jones) and electrostatic (e.g., Coulombic) field energies are calculated at each grid point using probe atoms. These thousands of energy values serve as the descriptors. Partial Least Squares (PLS) regression is then commonly used to correlate these descriptors with the biological activity, reducing dimensionality and mitigating the risk of overfitting [61].
Validation Workflow: The model is first validated internally using leave-one-out (LOO) cross-validation to calculate q². Subsequently, the final model, built using the optimal number of components from the cross-validation, is used to predict the activities of the external test set to calculate r²pred [60] [61].
The following diagram visualizes the relationship between alignment quality and the resulting validation metrics.
Table 2: Key Research Reagents and Computational Tools for 3D-QSAR
| Item/Resource | Function/Description | Example Use in 3D-QSAR |
|---|---|---|
| Molecular Database (e.g., ChEMBL) | A curated database of bioactive molecules with drug-like properties. | Source of experimental bioactivity data (e.g., IC₅₀, Ki) for model training and validation [50]. |
| Computational Chemistry Software (e.g., Sybyl-X, Forge, Schrodinger Suite) | Integrated platforms for molecular modeling, dynamics, and QSAR. | Used for energy minimization, conformational analysis, molecular alignment, descriptor calculation, and PLS regression [12] [14]. |
| Molecular Descriptors | Numerical representations of molecular structures and properties. | 3D fields (steric, electrostatic) in CoMFA/CoMSIA; quantum mechanical electrostatic potentials (ESP) as advanced descriptors [62]. |
| Validation Scripts/Functions | Custom or built-in code for calculating q², r², r²pred, and other metrics. | Automated calculation of statistical parameters post-modeling to objectively assess robustness and predictive power [59]. |
| Machine Learning Algorithms (e.g., ANN, Random Forest) | Non-linear algorithms for finding complex structure-activity relationships. | Used as alternative or complementary methods to traditional PLS for building highly predictive models, especially with quantum descriptors [63] [62]. |
The evaluation of 3D-QSAR models demands a multi-faceted approach that looks beyond a single statistical metric. The following key takeaways emerge from the comparative analysis of current research:
- q² is a Gatekeeper, Not a Guarantee: A high leave-one-out cross-validated R² (q²) is a necessary initial hurdle, but it is not a sufficient indicator of a model's predictive power. It should never be used in isolation [60].
- External Validation is Non-Negotiable: The predictive R² for an external test set (r²pred) is the gold standard for evaluating model utility and must be reported in any rigorous 3D-QSAR study [59] [60].
- Alignment is Foundational: The quality of molecular alignment is the single most critical factor influencing the signal in a 3D-QSAR model. Investing in a rigorous, unbiased alignment protocol before model building is paramount to success [14] [62].
- Embrace Advanced Methods: The integration of quantum mechanical descriptors, artificial neural networks, and sophisticated pairwise alignment techniques can significantly enhance predictive performance, as demonstrated in state-of-the-art studies on targets like the hERG channel [62].
By adhering to these principles and critically evaluating all key statistical metrics—q², r², and especially r²pred—within the context of a sound molecular alignment strategy, researchers can develop 3D-QSAR models that are not only statistically robust but also genuinely predictive, thereby accelerating the drug discovery process.
Molecular alignment is a critical and challenging step in three-dimensional quantitative structure-activity relationship (3D-QSAR) modeling. The process of superimposing molecules in a shared 3D space directly influences the calculation of molecular descriptors and, consequently, the quality and predictive power of the resulting models [17] [2]. For years, manual alignment based on a researcher's intuition and experience was the predominant method. However, this approach is subjective and time-consuming [17]. This guide provides an objective comparison between manual and automated alignment methodologies, evaluating their performance across various literature datasets to inform best practices for researchers in computational chemistry and drug development.
Performance Comparison: Quantitative Analysis
The table below summarizes key experimental findings from comparative studies that directly tested both manual and automated alignment methods on established datasets.
Table 1: Performance Comparison of Manual vs. Automated Alignment in 3D-QSAR Studies
| Dataset | Alignment Method | QSAR Method | Statistical Results (q² / r² / r²pred) |
Key Findings | Source |
|---|---|---|---|---|---|
| Cyclic Urea HIV-1 PR Inhibitors (n=113) | Manual | CoMFA/CoMSIA | Best q²: 0.649 |
Manual alignment yielded statistically higher internal validation values. | [3] |
| Automated (Docking) | CoMFA/CoMSIA | Best Predictive r²: 0.754 |
Automated alignment produced more robust models for external prediction. | [3] | |
| Literature Datasets (e.g., Steroids) | Manual | CoMFA/CoMSIA | N/S (Broadly comparable) | Manual alignment requires significant effort and introduces subjectivity. | [17] |
| Automated (FBSS) | CoMFA/CoMSIA | N/S (Broadly comparable) | FBSS generated predictive models automatically, saving time and effort. | [17] | |
| hERG Blockers (Subset 2, MW 301-350) | Automated (AlphaQ) | 3D-QSAR (ANN) | R²train: 0.98, R²test: 0.79 |
Quantum-mechanical alignment handled structurally diverse molecules effectively. | [62] |
Abbreviations: q²: Cross-validated correlation coefficient; r²: Non-cross-validated correlation coefficient; r²pred: Predictive correlation coefficient for an external test set; CoMFA: Comparative Molecular Field Analysis; CoMSIA: Comparative Molecular Similarity Indices Analysis; FBSS: Field-Based Similarity Searching; ANN: Artificial Neural Network; N/S: Not Specified in detail.
Key Performance Insights
- Predictive Robustness vs. Internal Fit: A study on HIV-1 protease inhibitors found that while manual alignment could produce models with a superior internal
q²(0.649), the models from automated (docked) alignments demonstrated higher predictiver²(0.754) for an external test set, indicating greater robustness and real-world applicability [3]. - Automation as a Complementary Tool: Research on FBSS automated alignment showed that the resulting QSAR models were "broadly comparable" in predictive performance to those from manual alignments. The authors position automation not as a replacement, but as a powerful complementary tool for initial screening and to suggest non-obvious alignments [17].
- Handling Structural Diversity: Advanced automated methods like the
AlphaQprotocol, which uses quantum mechanical cross-correlation for alignment, have proven effective for datasets with high structural diversity and varying molecular weights, achieving high predictive accuracy (R²testup to 0.79) without a common molecular scaffold [62].
Experimental Protocols in Literature
Manual Alignment Workflow
The manual alignment protocol is often the benchmark against which new methods are measured. It is a multi-step process that relies heavily on researcher expertise [2].
- Data Preparation: A dataset of compounds with experimentally determined biological activities (e.g., IC₅₀) is assembled. The initial 2D structures are converted into 3D models and geometry-optimized using force fields (e.g., UFF) or quantum mechanical methods to achieve low-energy, realistic conformations [2].
- Identification of a Common Framework: The researcher identifies a common structural feature across all molecules. This could be a rigid ring template, a maximum common substructure (MCS), or an obvious pattern of pharmacophore points (e.g., hydrogen bond donors/acceptors, hydrophobic regions) [17] [2].
- Superposition: Using molecular visualization software, each molecule is manually rotated and translated to fit its common framework onto that of a chosen reference molecule, typically the most active compound or a known crystallographic ligand [17] [2].
- Validation: The quality of the alignment is visually inspected and judged based on the researcher's experience. This step is inherently subjective, and different researchers may produce different alignments for the same dataset [17].
Automated Alignment Workflows
Automated methods aim to remove subjectivity and reduce manual effort. The search results highlight several distinct approaches.
Field-Based Similarity Searching (FBSS)
This method aligns molecules based on the similarity of their molecular fields rather than atom positions [17].
- Field Calculation: Each molecule is positioned at the center of a 3D grid. Molecular field values, such as the steric (van der Waals) and electrostatic (Coulombic) potentials, are calculated at each grid point [17].
- Similarity Maximization: The FBSS algorithm automatically rotates and translates each molecule to maximize the similarity between its molecular fields and those of a reference molecule. This is typically quantified using a similarity index, such as the Carbo index [17].
- Alignment Output: The result is a set of aligned molecules ready for descriptor calculation in methods like CoMFA or CoMSIA [17].
Molecular Docking-Based Alignment
This structure-based method uses the known 3D structure of the target protein to guide alignment [3].
- Protein Preparation: The 3D structure of the target protein (e.g., from X-ray crystallography) is prepared by adding hydrogen atoms, assigning charges, and defining the active site.
- Ligand Docking: Each flexible ligand is computationally docked into the protein's active site. The docking algorithm scores and ranks different poses based on the predicted binding affinity [3].
- Pose Extraction: The highest-ranked binding pose for each ligand is extracted. The alignment of these poses relative to the protein defines their relative alignment to each other, which is then used for 3D-QSAR [3].
Advanced Algorithmic Alignment (AlphaQ)
For structurally diverse datasets lacking a common core, advanced methods like AlphaQ are employed [62].
- Subset Division: The full dataset is divided into subsets based on molecular properties, such as molecular weight, to ensure alignment relevance [62].
- Pairwise Quantum-Mechanical Alignment: Within each subset, molecules are aligned pairwise to a template. The alignment is optimized by maximizing the quantum mechanical cross-correlation of their electrostatic potentials, rather than relying on atom-by-atom matching [62].
- Descriptor Calculation and Modeling: The optimally aligned structures are used to calculate quantum mechanical descriptors (e.g., electrostatic potential), which are then fed into a machine learning algorithm like an Artificial Neural Network (ANN) to build the QSAR model [62].
The following diagram illustrates the logical flow and key decision points for selecting an alignment methodology.
The Scientist's Toolkit: Essential Research Reagents & Software
The table below catalogs key computational tools and methodologies referenced in the comparative literature.
Table 2: Key Research Tools for Molecular Alignment and 3D-QSAR
| Tool / Method | Type | Primary Function in Alignment/QSAR | Relevant Citation |
|---|---|---|---|
| Sybyl (Tripos) | Software Platform | Classic proprietary software for manual alignment, CoMFA, and CoMSIA modeling. | [17] [55] |
| FBSS (Field-Based Similarity Searching) | Algorithm | Automated alignment by maximizing the similarity of molecular fields (steric, electrostatic). | [17] |
| Molecular Docking | Computational Method | Generates alignments by predicting the binding pose of ligands within a protein's active site. | [3] |
| AlphaQ | Algorithm | Aligns diverse molecules by optimizing quantum mechanical cross-correlation of electrostatic potentials. | [62] |
| Py-CoMSIA | Software Library | An open-source Python implementation of CoMSIA, increasing accessibility to 3D-QSAR methodologies. | [55] |
| CoMFA | 3D-QSAR Method | Requires aligned molecules to calculate steric and electrostatic interaction fields on a 3D grid. | [17] [2] |
| CoMSIA | 3D-QSAR Method | Requires aligned molecules to calculate similarity indices for steric, electrostatic, hydrophobic, and H-bond fields. It is generally less sensitive to minor alignment deviations than CoMFA. | [41] [2] [55] |
The choice between manual and automated alignment is not a simple binary decision. Evidence from literature shows that manual alignment can produce models with excellent internal validation metrics, but it is subjective and labor-intensive. Conversely, automated methods offer objectivity, reproducibility, and scalability, with studies demonstrating that they can produce models of comparable, and sometimes superior, predictive robustness for external compounds [17] [3]. Advanced automated techniques like AlphaQ and FBSS are particularly valuable for handling structurally diverse datasets where manual alignment is most challenging [17] [62].
The most effective strategy is often a hybrid one. Automated alignments can serve as an excellent starting point, providing an objective baseline or suggesting novel superposition hypotheses that a researcher can then refine based on their biochemical intuition and knowledge of the target. The growing development of open-source tools, such as Py-CoMSIA, further democratizes access to these advanced computational methods, empowering more researchers to incorporate robust 3D-QSAR into their drug discovery pipelines [55].
Interpreting contour maps transcends mere statistical analysis in 3D-QSAR; it represents the crucial bridge between computational models and actionable biological insights. While statistical metrics validate model robustness, true scientific advancement emerges from interpreting the steric and electrostatic fields depicted in contour maps to understand ligand-receptor interactions. This process faces a fundamental challenge: contour maps are highly dependent on the molecular alignment method used to generate them. Different alignment strategies can produce dramatically different contour visualizations, potentially leading to conflicting structural interpretations and pharmacological hypotheses. This guide provides an objective comparison of predominant molecular alignment techniques, evaluating their performance in generating reliable, interpretable contour maps for 3D-QSAR research. We focus specifically on how alignment choices impact the contour maps that scientists use to guide molecular design, supported by experimental data from structured comparative studies.
Comparative Analysis of Molecular Alignment Methods
The choice of molecular alignment strategy directly influences the contour maps generated in 3D-QSAR studies, with significant implications for model interpretation and predictive accuracy. The following analysis compares the predominant methodologies.
Performance Comparison of Alignment Methods
Table 1: Quantitative Comparison of Molecular Alignment Methods for 3D-QSAR
| Alignment Method | Average R²Test (Androgen Receptor Dataset) | Computational Efficiency | Required Expertise Level | Contour Map Reproducibility | Key Strengths | Major Limitations |
|---|---|---|---|---|---|---|
| 2D->3D Conversion (No Alignment) [10] | 0.61 | Very High | Low | Moderate | Speed, suitability for large datasets; avoids alignment subjectivity [10]. | Conformations may not be biologically relevant; potential for misleading contours. |
| Global Energy Minimization [10] | 0.56 - 0.61 | Low | Medium | High | Physically realistic conformations; high reproducibility [10]. | Computationally intensive; biologically active conformation not guaranteed. |
| Template-Based Alignment [10] | 0.56 - 0.61 | Very Low | High | Low | Potentially biologically relevant alignment; uses known pharmacophore information [10]. | Highly subjective; choice of template biases results; low reproducibility. |
| Consensus Modeling (Aggregate) [10] | 0.65 | Lowest | High | Variable | Highest predictive accuracy; mitigates bias from any single method [10]. | Maximizes computational cost and complexity; interpretation can be challenging. |
Interpretation of Comparative Data
The experimental data, derived from a diverse dataset of 146 androgen receptor binders, reveals critical insights for practitioners [10]. Counterintuitively, the simplest method—2D->3D conversion—achieved a predictive accuracy superior to more computationally intensive strategies, producing an R²Test of 0.61 in only 3-7% of the time required by other methods [10]. This suggests that for certain receptor targets, particularly those like the androgen receptor where highly active ligands are fairly inflexible, exhaustive conformational analysis may be unnecessary.
However, the consensus approach, which aggregates predictions from models built on different conformations, achieved the highest overall accuracy (R²Test = 0.65) [10]. This demonstrates that while a single simple conformation can be effective, integrating multiple alignment perspectives can capture complementary aspects of the ligand-receptor interaction, leading to more robust models and, consequently, more reliable contour maps for interpretation.
Experimental Protocols for Method Comparison
To ensure reproducible and meaningful comparisons of alignment methods, researchers should adhere to standardized experimental protocols.
Dataset Curation and Preparation
The foundation of any robust comparison is a well-curated dataset. The referenced study utilized 146 compounds with known binding affinities (Relative Binding Affinity, RBA) to the androgen receptor, sourced from the NCTR Endocrine Disruption Knowledge Base (EDKB) [10]. Key criteria include:
- Structural Diversity: The dataset should encompass multiple chemical classes (e.g., steroids, DESs, phenols, pesticides) to test the generality of the alignment method [10].
- Measured Affinity: All biological data (e.g., IC₅₀, Ki) should ideally be generated from a single laboratory using consistent protocols to minimize experimental noise [10].
- Data Transformation: Affinity data is often transformed to a logarithmic scale (e.g., log(RBA)) to improve normality for statistical modeling [10].
Conformational Generation and Alignment Workflow
Each alignment method follows a distinct computational pathway.
Protocol for Template-Based Alignment:
- Template Selection: Choose one or more high-affinity, rigid molecules believed to represent the key pharmacophore.
- Alignment Execution: Using molecular modeling software (e.g., with force fields considering equal electronic and steric contributions or "Best-for-Each" template settings), flexibly align each molecule in the dataset to the template(s) [10].
- Conformation Extraction: Output the aligned conformation of each molecule for subsequent 3D-QSAR analysis.
Protocol for Global Energy Minimization:
- Conformational Search: Perform a systematic search or use stochastic methods to explore the potential energy surface (PES) of each molecule.
- Energy Optimization: Identify the global minimum energy conformation using semi-empirical or quantum mechanical (QM) methods for precise determination [10].
- Structure Output: Use the optimized global minimum conformation for model building.
Protocol for 2D->3D Conversion:
- Structure Import: Directly convert 2D structural representations (e.g., SMILES strings) into 3D coordinates using molecular mechanics as implemented in tools like Jmol or by importing from databases like ChemSpider [10].
- No Further Optimization: Use these non-energy-optimized, non-aligned 3D structures directly in descriptor calculation and model building [10].
Model Building and Contour Map Generation
Following alignment, a standardized 3D-QSAR workflow is applied:
- Descriptor Calculation: Place all aligned molecules into a common 3D grid. Calculate interaction energy descriptors (e.g., steric, electrostatic) at each grid point.
- Model Training: Use Partial Least Squares (PLS) regression to build a statistical model linking the 3D descriptors to biological activity [10].
- Contour Map Generation: Visualize the model coefficients as 3D contour maps. These maps highlight regions in space where specific molecular properties (e.g., increased steric bulk, positive electrostatic potential) are associated with enhanced or diminished biological activity.
Figure 1: Experimental workflow for comparing molecular alignment methods in 3D-QSAR.
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 2: Key Research Reagents and Computational Tools for 3D-QSAR Alignment Studies
| Item Name | Function/Description | Relevance to Contour Interpretation |
|---|---|---|
| Diverse Chemical Dataset | A set of molecules with known biological activity and structural diversity. | The foundational input; ensures models and contours are generalizable and not based on a narrow chemical space [10]. |
| Molecular Mechanics Software (e.g., Jmol) | Performs 2D->3D conversion and basic energy minimization using classical force fields. | Generates initial 3D coordinates quickly, forming the baseline for method comparison [10]. |
| Quantum Mechanical (QM) Software | Provides high-accuracy quantum mechanical calculations for determining global energy minima. | Produces theoretically rigorous, energy-optimized conformations for "gold standard" comparison [10]. |
| 3D-QSAR Software Platform (e.g., for CoMFA/CoMSIA) | Provides the computational environment for descriptor calculation, PLS regression, and contour map visualization. | The primary tool for translating aligned molecular sets into interpretable 3D contour maps. |
| Structural Database (e.g., ChemSpider) | A source for 2D and 3D structural information of molecules. | Provides readily available 2D->3D structures for testing the non-aligned conformation approach [10]. |
| Template Molecules | Known active, rigid molecules used as references for alignment. | Critical for the template-based alignment method; their selection heavily influences the resulting contours [10]. |
Interpreting contour maps to extract meaningful SAR insights is profoundly dependent on the underlying molecular alignment strategy. The experimental data demonstrates that there is no single "best" method universally; the optimal choice is context-dependent. For rapid screening of large, diverse datasets or for modeling receptors with relatively inflexible ligands, the 2D->3D approach offers an unexpectedly powerful and efficient path to interpretable contours [10]. When resource-intensive modeling is feasible and the highest predictive accuracy is paramount, a consensus approach that aggregates multiple alignment strategies provides the most robust and reliable contour maps for guiding molecular design [10]. Ultimately, scientists must weigh the trade-offs between computational cost, theoretical justification, and empirical performance to select the alignment method that best illuminates the structure-activity relationships central to their research.
In the landscape of modern drug discovery, lead optimization represents a critical phase where promising chemical compounds are refined into viable preclinical candidates. The core challenge lies in enhancing a molecule's efficacy and pharmacokinetic properties while minimizing its toxicity, a process that requires the meticulous exploration of vast chemical space [64] [65]. Within this domain, three-dimensional quantitative structure-activity relationship (3D-QSAR) modeling has emerged as a pivotal computational technique, enabling researchers to correlate the biological activity of compounds with their three-dimensional molecular fields [41]. The predictive power and ultimate success of any 3D-QSAR model are fundamentally dependent on a single, crucial step: the molecular alignment of the compounds under study. The choice of alignment method—whether manual, based on known pharmacophore elements or crystal structures, or automated, driven by molecular docking—can significantly influence the model's quality and its reliability in prospective drug design [3].
This guide objectively compares manual and automated alignment methods within 3D-QSAR, framing the discussion within the broader thesis that automated techniques can produce models of comparable, and sometimes superior, predictiveness to traditional manual approaches. We will provide a detailed comparison of their performance, supported by experimental data and case studies, and outline the essential toolkit required for implementation.
Comparative Analysis of Alignment Methods in 3D-QSAR
Molecular alignment in 3D-QSAR involves superimposing a set of molecules to maximize the similarity of their steric and electrostatic fields in a hypothesized active orientation. The two predominant paradigms for achieving this alignment are manual and automated methods, each with distinct philosophies and practical implications.
- Manual Alignment: This traditional approach relies heavily on researcher intuition and existing structural biology data. The alignment is typically guided by a common pharmacophore, the crystallographically determined binding mode of a lead compound, or the docking of a rigid, active compound to define a template conformation [3]. The process is iterative and expert-dependent, leveraging deep knowledge of the target's active site and ligand-receptor interactions.
- Automated Alignment: This method reduces subjective bias by using computational algorithms to generate the molecular superposition. A common automated approach involves molecular docking, where each ligand is independently docked into the target's binding site, and the resulting poses are used for the 3D-QSAR alignment [3]. This technique is particularly valuable when the structure of the target protein is well-characterized, as it directly incorporates information about the binding pocket.
A seminal study provides a direct, quantitative comparison of these two approaches, offering critical insights for the thesis that automated methods are robust and reliable [3]. The research utilized a set of 113 flexible cyclic urea inhibitors of human immunodeficiency virus protease (HIV-1 PR) to build both CoMFA and CoMSIA models.
Table 1: Quantitative Comparison of Manual vs. Automated Alignment for 3D-QSAR Model Quality [3]
| Alignment Method | 3D-QSAR Technique | Cross-Validated R² (q²) | Predictive R² for External Test Set | Model Robustness & Generalizability |
|---|---|---|---|---|
| Manual Alignment | CoMFA / CoMSIA | Statistically higher values (e.g., best q² = 0.649) | Lower predictive power on external set | More tailored to the training set |
| Automated Alignment (Molecular Docking) | CoMFA / CoMSIA | Slightly lower but statistically significant | Higher predictive power (e.g., predictive r² = 0.754) | More robust for predicting new chemotypes |
The data in Table 1 reveals a critical distinction: while manual alignment can produce models with slightly superior internal validation metrics (e.g., q²), the models derived from automated docking alignment demonstrated greater predictive power and robustness when applied to an external inhibitor set [3]. Furthermore, both models identified similar key interactions with amino acid residues in the HIV-1 PR active site (e.g., hydrogen bonds with Gly48 and Asp30), validating the automated method's ability to capture biologically relevant features [3].
Another comparative study on arylbenzofuran histamine H3 receptor antagonists found that traditional 2D methods like Multiple Linear Regression (MLR) and Artificial Neural Networks (ANN) could perform on par with or even better than 3D-QSAR methods like HASL in predicting binding affinities [52]. This underscores that the choice of modeling technique itself is context-dependent, but for 3D-QSAR, automated alignment is a validated and often preferable strategy.
Experimental Protocols for 3D-QSAR Model Building and Validation
To ensure the development of predictive and reliable 3D-QSAR models, a rigorous and standardized experimental protocol must be followed. The following workflow delineates the key stages, from initial data preparation through to final model application, and can be adapted for either manual or automated alignment.
The following diagram illustrates the comprehensive workflow for building and validating a 3D-QSAR model.
Detailed Methodologies
Data Set Preparation and Conformational Analysis
The foundation of a robust QSAR model is a high-quality, curated dataset. The process begins with the selection of a congeneric series of compounds with a consistent mechanism of action and reliably measured biological activity data (e.g., IC₅₀, Kᵢ) [41]. For each compound, energy minimization and a systematic conformational search are performed using molecular mechanics or quantum chemical methods to identify low-energy conformers. Software tools like Sybyl-X are commonly used for these construction and optimization steps [41]. The most relevant bioactive conformer is typically selected as a template for subsequent alignment.
Molecular Alignment Procedures
This is the critical step where manual and automated paths diverge.
Manual Alignment Protocol: Researchers superimpose molecules based on a common structural framework or pharmacophore hypothesis. This often uses the crystallographically determined binding mode of a high-affinity ligand from a protein-ligand complex as a fixed template [3]. The alignment is manually refined to maximize the overlap of key functional groups and the molecular volume considered essential for binding.
Automated Alignment Protocol: This method leverages computational docking to define alignment. Each molecule is independently docked into the target protein's binding site using a program like Glide [66]. The docking poses are carefully analyzed, and a consensus binding mode is identified. The ligands are then aligned based on these docked conformations, ensuring the superposition reflects their proposed orientation within the binding pocket [3]. This process is less subjective and directly incorporates the 3D structure of the target.
3D-QSAR Model Calculation and Validation
Once aligned, the molecular set is used to calculate 3D fields. In techniques like Comparative Molecular Similarity Indices Analysis (CoMSIA), steric, electrostatic, hydrophobic, and hydrogen-bond donor/acceptor fields are commonly computed [41]. The dataset is split into a training set (typically ~80%) for model building and a test set (~20%) for external validation. The model is constructed using partial least squares (PLS) regression. The COMSIA model from a recent study on MAO-B inhibitors, for instance, showed high internal consistency (r² = 0.915) and a cross-validated coefficient (q²) of 0.569, indicating good predictive ability [41].
Validation is a non-negotiable step. It involves:
- Internal Validation: Using the training set with cross-validation techniques (e.g., leave-one-out) to generate the q² value [41] [3].
- External Validation: Predicting the activity of the withheld test set to calculate the predictive r², which is the true measure of a model's utility [3].
- Statistical Scrutiny: Analysis of standard error of estimate (SEE) and F-value to assess model significance [41].
Case Studies: Prospective Validation in Lead Optimization
The true test of any computational model is its successful application in the prospective design of novel, potent compounds. The following case studies demonstrate this validation, showcasing how 3D-QSAR models, particularly those leveraging modern alignment and simulation techniques, directly contribute to successful lead optimization.
Case Study 1: Designing Novel MAO-B Inhibitors for Neuroprotection
A 2025 study aimed to develop novel neuroprotective agents by inhibiting Monoamine Oxidase B (MAO-B), a target for Parkinson's and Alzheimer's disease [41]. Researchers built a 3D-QSAR model using the CoMSIA method on a series of 6-hydroxybenzothiazole-2-carboxamide derivatives.
- Experimental Protocol: Compounds were constructed and optimized using ChemDraw and Sybyl-X software. The molecular alignment was likely guided by the shared benzothiazole core and established pharmacophore points. The resulting CoMSIA model exhibited excellent statistical quality (q² = 0.569, r² = 0.915), providing a reliable predictive tool [41].
- Prospective Application & Validation: Leveraging the model's contour maps, the team designed a series of novel derivatives, predicting their IC₅₀ values in silico. Among them, compound 31.j3 was predicted to be highly potent. This prediction was validated through molecular docking, where 31.j3 achieved a high score, and crucially, by molecular dynamics (MD) simulations [41]. The MD simulations confirmed the stable binding of 31.j3 to the MAO-B receptor, with RMSD values fluctuating minimally between 1.0 and 2.0 Å, indicating a stable complex. Energy decomposition analysis further detailed the key van der Waals and electrostatic interactions stabilizing the bond [41]. This end-to-end workflow—from QSAR design to dynamics validation—exemplifies a successful prospective application in lead optimization.
Case Study 2: Advancing HIV-1 Protease Inhibitors with Automated Alignment
The previously cited study on HIV-1 protease inhibitors provides compelling evidence for the use of automated alignment in a prospective context [3]. The research established that 3D-QSAR models built using automated (docking-based) alignment were more robust and predictive for external compounds than those from manual alignment.
- Experimental Protocol: A set of 113 cyclic urea inhibitors was aligned both manually and via automated molecular docking. CoMFA and CoMSIA models were developed from both alignments [3].
- Prospective Application & Validation: The key finding was that the automated-alignment model was better at predicting the activities of a true external test set. This superior predictive r² (0.754) directly translates to higher confidence in prospective drug design. A research team could use such a model to screen virtual libraries or guide the optimization of newly synthesized compounds with a greater assurance that the predicted activities are accurate, thereby reducing the number of cycles needed in the "design-make-test-analyze" loop [3].
Case Study 3: Integrating FEP and 3D-QSAR in a Multi-Tool Workflow
Beyond pure 3D-QSAR, the most powerful contemporary applications involve its integration with more advanced physics-based methods. A review in Accounts of Chemical Research highlighted a workflow for discovering non-nucleoside inhibitors of HIV reverse transcriptase (NNRTIs) that combines de novo design (using BOMB), virtual screening (using Glide), and lead optimization driven by Free Energy Perturbation (FEP) calculations [66].
- Experimental Protocol: Initial leads were generated and optimized using a combination of docking and QSAR-like scoring. The most promising candidates were then subjected to FEP calculations, which provide highly accurate binding affinity predictions by computationally "transforming" one ligand into another within the simulated binding site [66] [67].
- Prospective Application & Validation: This FEP-guided optimization was instrumental in advancing initial leads with low-µM activity to highly potent inhibitors with low-nM activity [66]. This case demonstrates that 3D-QSAR and machine learning models are excellent for rapid screening and idea prioritization, while FEP can be applied as a high-precision tool for final optimization, forming a powerful multi-tiered computational strategy for lead optimization.
The Scientist's Toolkit: Essential Research Reagents and Solutions
Implementing the experimental protocols described requires a suite of specialized software tools and reagents. The following table details key solutions essential for 3D-QSAR and prospective drug design.
Table 2: Essential Research Reagent Solutions for 3D-QSAR and Lead Optimization
| Tool / Solution Name | Primary Function | Key Application in Workflow |
|---|---|---|
| Sybyl-X [41] | Molecular modeling and QSAR | Compound construction, energy minimization, conformational analysis, and CoMSIA model calculation. |
| ChemDraw [41] | Chemical structure drawing | Initial 2D structure depiction and preparation for 3D model conversion. |
| Glide [66] | Molecular docking | Virtual screening of compound libraries and generation of poses for automated molecular alignment. |
| BOMB (Biochemical and Organic Model Builder) [66] | De novo ligand design | Growing molecules by adding substituents to a molecular core for lead generation and optimization. |
| Orion 3D-QSAR [68] | Machine Learning-based 3D-QSAR | Building predictive QSAR models featurized with 3D shape and electrostatics; provides prediction confidence estimates. |
| Octet BLI Systems [69] | Label-free binding affinity measurement | Experimental validation of binding kinetics (ka, kd, KD) and affinity ranking during lead optimization. |
| Molecular Dynamics (MD) Software (e.g., GROMACS, AMBER) | Simulating biomolecular systems | Assessing binding stability and dynamic behavior of protein-ligand complexes, as used in the MAO-B case study [41]. |
| Free Energy Perturbation (FEP) Software [66] [67] | High-accuracy binding affinity prediction | Prioritizing design ideas for synthesis based on precise affinity calculations during late-stage lead optimization. |
The comparative analysis and case studies presented validate a central thesis in modern computational drug discovery: automated alignment methods for 3D-QSAR, particularly those based on molecular docking, can yield models that are not only statistically sound but also highly predictive and robust for prospective compound design [3]. While manual alignment retains value in specific scenarios, the data-driven, less subjective nature of automated alignment makes it a powerful and often preferable approach, especially when a high-resolution protein structure is available.
The future of lead optimization lies in the intelligent integration of these methods. As evidenced by the most successful case studies, 3D-QSAR acts as an efficient engine for generating and prioritizing design ideas. Its impact is magnified when used in concert with high-precision tools like FEP for final affinity optimization [66] [67] and molecular dynamics for validating binding stability [41]. Furthermore, the advent of machine learning in 3D-QSAR, which uses shape and electrostatic featurizations, promises even greater predictive power and the valuable ability to estimate prediction confidence [68]. For researchers and scientists, this evolving multi-method toolkit provides an unprecedented capacity to navigate the challenges of lead optimization, systematically transforming promising leads into effective and safe clinical candidates.
In modern computational drug discovery, the integration of three-dimensional quantitative structure-activity relationship (3D-QSAR) modeling with molecular docking has become a cornerstone methodology for rational drug design. While each technique provides valuable standalone insights, their combination creates a synergistic workflow that significantly enhances the efficiency and predictive power of virtual screening campaigns. This integrated approach allows researchers to not only understand the key structural features governing biological activity but also visualize how these features facilitate molecular recognition at the target binding site.
The fundamental synergy arises from the complementary strengths of each method. 3D-QSAR techniques, particularly Comparative Molecular Similarity Indices Analysis (CoMSIA) and Comparative Molecular Field Analysis (CoMFA), generate contour maps that highlight regions where specific molecular properties (steric, electrostatic, hydrophobic, hydrogen bond donor/acceptor) enhance or diminish biological activity [16]. Molecular docking then provides the structural context for these observations by predicting the binding orientation and key interactions between ligands and their target proteins [70]. When used iteratively, this combination enables a powerful feedback loop where docking validates QSAR predictions, and QSAR guides the optimization of docked compounds.
Key Methodological Components
3D-QSAR Techniques: CoMFA and CoMSIA
Comparative Molecular Field Analysis (CoMFA) represents one of the most established 3D-QSAR approaches, calculating steric and electrostatic fields around aligned molecules using a Lennard-Jones and Coulomb potential, respectively [16]. The model results in contour maps that identify regions where steric bulk or specific electrostatic properties are favorable or unfavorable for activity.
Comparative Molecular Similarity Indices Analysis (CoMSIA) extends beyond CoMFA by incorporating additional molecular fields and addressing some limitations of the original approach. Unlike CoMFA's Lennard-Jones and Coulomb potentials, CoMSIA employs a Gaussian function to calculate similarity indices, producing more interpretable contour maps without abrupt field changes [16]. CoMSIA typically evaluates five distinct property fields:
- Steric (S) and Electrostatic (E) fields analogous to CoMFA
- Hydrophobic (H) field representing desolvation effects
- Hydrogen bond donor (D) and acceptor (A) fields characterizing specific interactions
The statistical quality of 3D-QSAR models is evaluated through multiple parameters, including cross-validated correlation coefficient (Q²), conventional correlation coefficient (R²), standard error of estimate (SEE), and predictive R² (R²pred) for external test sets [12] [71]. A Q² value > 0.5 is generally considered indicative of a robust model with good predictive capability [71].
Molecular Docking Fundamentals
Molecular docking computationally predicts the optimal binding orientation and conformation of a small molecule ligand within a protein's binding site [70]. The process involves two main components: conformational sampling of the ligand in the binding site and scoring function evaluation to rank the predicted poses based on estimated binding affinity.
Docking provides atomic-level insights into protein-ligand interactions, identifying specific:
- Hydrogen bonds and their geometries
- Hydrophobic interactions and π-π stacking
- Electrostatic interactions and salt bridges
- Steric complementarity between ligand and binding site
The binding affinity scores (typically in kcal/mol) allow for rapid virtual screening of compound libraries and prioritization of candidates for further investigation [72].
Integrated Workflow and Experimental Protocols
The synergistic application of 3D-QSAR and molecular docking follows a systematic workflow that maximizes the strengths of both approaches. This integrated methodology has been successfully applied across multiple therapeutic areas, from neurodegenerative disorders to oncology and metabolic diseases.
Standard Experimental Protocol
The integrated workflow follows these key experimental stages:
Dataset Curation and Preparation: A series of compounds with known biological activities (IC₅₀ or Kᵢ values) is collected from literature or experimental data. The biological activities are typically converted to pIC₅₀ values [-log₁₀(IC₅₀)] for QSAR analysis [73]. The dataset is divided into training (typically 75-80%) and test sets (20-25%) for model development and validation [74] [71].
Molecular Alignment: Proper alignment of molecules is the most critical step in 3D-QSAR model development. Common approaches include:
3D-QSAR Model Development: CoMFA and CoMSIA models are built using the training set compounds. Partial Least Squares (PLS) regression correlates the field descriptors with biological activity. Leave-One-Out (LOO) cross-validation determines the optimal number of components and Q² value [71].
Model Validation: Both internal (cross-validation) and external (test set prediction) validations are performed. A robust model should have Q² > 0.5 and R²pred > 0.6 [71]. The contour maps are analyzed to identify key structural requirements for activity.
Compound Design and Prioritization: New compounds are designed based on contour map insights. Molecular docking screens these designed compounds to evaluate binding modes and interactions with key amino acid residues [12] [75].
Binding Stability Assessment: Molecular dynamics simulations (typically 50-100 ns) assess the stability of protein-ligand complexes and validate docking predictions [12] [70] [72].
ADMET Profiling: Absorption, distribution, metabolism, excretion, and toxicity properties are predicted in silico to evaluate drug-likeness and prioritize candidates for synthesis [70] [71].
Key Research Reagents and Computational Tools
Table 1: Essential Research Reagents and Software Solutions for Integrated 3D-QSAR and Docking Studies
| Category | Specific Tools/Reagents | Function/Purpose | Application Examples |
|---|---|---|---|
| Molecular Modeling Suites | SYBYL/X, Schrödinger, MOE | Comprehensive platforms for 3D-QSAR, molecular docking, and simulation | CoMFA/CoMSIA model development [12] [71] |
| Open-Source QSAR Tools | Py-CoMSIA (Python) | Open-source implementation of CoMSIA methodology | Accessible 3D-QSAR without proprietary software [16] |
| Docking Software | AutoDock Vina, GOLD | Molecular docking and virtual screening | Binding pose prediction and affinity ranking [71] |
| Dynamics Packages | GROMACS, AMBER | Molecular dynamics simulations | Binding stability assessment (50-100 ns) [70] [73] |
| Protein Data Sources | RCSB PDB | Experimentally-determined protein structures | Source of target structures for docking [74] |
| Compound Databases | PubChem, ZINC | Libraries of available compounds | Virtual screening and similarity searching [73] |
Case Studies and Comparative Performance
The integrated 3D-QSAR and docking approach has demonstrated significant success across multiple therapeutic areas. The following case studies highlight the performance and advantages of this synergistic methodology.
Table 2: Comparative Performance of Integrated 3D-QSAR and Docking Across Therapeutic Areas
| Therapeutic Area | Target Proteins | 3D-QSAR Statistics | Docking Performance | Key Findings | Reference |
|---|---|---|---|---|---|
| Neurodegenerative Diseases | MAO-B | CoMSIA: Q²=0.569, R²=0.915 | Improved binding scores vs. reference | Compound 31.j3 showed stable binding (RMSD 1.0-2.0Å) in MD simulations | [12] |
| Oncology (Breast Cancer) | CDK2, EGFR, Tubulin | CoMSIA/SEHDA: Q²=0.814, R²=0.967 | Binding affinity: -7.2 to -9.8 kcal/mol | Multi-target inhibitors with improved affinity over reference compounds | [72] |
| Diabetes | α-Glucosidase | CoMSIA: Q²=0.616, R²=0.928 | Stable binding in active site | Designed compounds M1, M2 showed promising anti-diabetic potential | [70] |
| Oncology (Prostate Cancer) | PLK1 | CoMFA: Q²=0.67, R²=0.992 | Interaction with key residues R136, R57, Y133 | Identified stable inhibitors (50ns MD) with potential for prostate cancer therapy | [71] |
| Neurological Disorders | GSK-3β | CoMFA: Q²=0.505, R²=0.935 | Strong binding to active site | Compounds 3X and 9X predicted with higher activity than lead compound | [75] |
MAO-B Inhibitors for Neurodegenerative Diseases
In the development of monoamine oxidase B (MAO-B) inhibitors for neurodegenerative diseases, researchers integrated CoMSIA-based 3D-QSAR with molecular docking and dynamics simulations. The established CoMSIA model demonstrated excellent statistical quality (Q² = 0.569, R² = 0.915), highlighting the importance of electrostatic and hydrophobic fields for MAO-B inhibition [12].
The contour maps informed the design of novel 6-hydroxybenzothiazole-2-carboxamide derivatives, with compound 31.j3 emerging as a promising candidate. Molecular docking revealed superior binding orientation and interactions compared to reference compounds. Subsequent molecular dynamics simulations confirmed the stability of the MAO-B-31.j3 complex, with RMSD values fluctuating between 1.0-2.0 Å over the simulation period, indicating excellent conformational stability [12]. Energy decomposition analysis further identified the key amino acid residues contributing to binding affinity through van der Waals and electrostatic interactions.
Multi-Target Cancer Therapy
In oncology drug discovery, researchers applied the integrated approach to develop 2-phenylindole derivatives as multi-target inhibitors for CDK2, EGFR, and tubulin simultaneously. The CoMSIA/SEHDA model exhibited exceptional predictive power (Q² = 0.814, R² = 0.967, R²pred = 0.722) [72].
Six newly designed compounds demonstrated improved binding affinities (-7.2 to -9.8 kcal/mol) across all three targets compared to reference molecules. Molecular docking revealed comprehensive interaction profiles with key residues in all target proteins. Molecular dynamics simulations confirmed the stability of these complexes throughout 100 ns trajectories, validating the multi-target inhibition strategy [72]. This approach successfully addressed the limitation of single-target therapies in cancer treatment, which often lead to drug resistance through compensatory pathway activation.
The integration of 3D-QSAR and molecular docking represents a powerful synergy in computational drug discovery, combining the quantitative predictive power of QSAR with the structural insights of molecular docking. This methodology enables researchers to understand not only what structural features enhance activity but also why these features are important based on the three-dimensional interaction with the biological target.
The case studies across therapeutic areas demonstrate that this integrated approach consistently yields compounds with improved binding affinity, optimized interactions, and favorable stability profiles. The iterative nature of the workflow—where docking validates QSAR predictions and QSAR guides compound optimization—creates a efficient design cycle that accelerates the drug discovery process.
As computational methods continue to evolve, particularly with the development of open-source tools like Py-CoMSIA that increase accessibility [16], the integration of 3D-QSAR with docking and molecular dynamics simulations is poised to become even more central to rational drug design. This synergistic methodology provides a robust framework for addressing the increasing complexity of drug targets and the need for more efficient discovery pipelines.
Conclusion
Molecular alignment is not merely a preliminary step but a decisive factor that shapes the predictive power and interpretability of 3D-QSAR models. This analysis demonstrates that no single alignment method is universally superior; the optimal choice is contingent on the dataset's structural homogeneity, the flexibility of the compounds, and the specific project goals, such as lead optimization versus scaffold hopping. The field is increasingly moving towards automated, field-based methods and alignment-independent descriptors to reduce subjectivity and enhance reproducibility, while the integration of 3D-QSAR with molecular docking and dynamics provides a more holistic view of ligand-receptor interactions. Looking forward, the incorporation of AI and machine learning promises to further revolutionize alignment strategies, enabling the navigation of vast chemical spaces with unprecedented efficiency. For biomedical research, mastering these comparative alignment techniques is paramount for accelerating the discovery of novel, effective therapeutics with improved safety profiles, ultimately bridging the gap between computational prediction and clinical success.
References
- 1. 3D-QSAR : Principles and Methods [https://drugdesign.org/chapters/3d-qsar/]
- 2. Beginner's Guide to 3D-QSAR in Drug Design [https://neovarsity.org/blogs/beginners-guide-3d-qsar-drug-design]
- 3. the quality and predictiveness between Comparing models... 3 D QSAR [https://pubmed.ncbi.nlm.nih.gov/15154745/]
- 4. Receptor-based 3D-QSAR in Drug Design [https://pubmed.ncbi.nlm.nih.gov/26369822/]
- 5. Docking, Interaction Fingerprint, and Three-Dimensional ... [https://www.frontiersin.org/journals/chemistry/articles/10.3389/fchem.2019.00496/full]
- 6. QSAR Toolbox [https://qsartoolbox.org/]
- 7. Molecular Interaction Fields Describing Halogen Bond ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11323840/]
- 8. Recent Developments in 3D QSAR and Molecular Docking ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7123761/]
- 9. An improved 3D quantitative structure-activity relationships ... [https://www.sciencedirect.com/science/article/pii/S2667318523000090]
- 10. Alignment-independent technique for 3D QSAR analysis - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4833814/]
- 11. A comparison between 2D and 3D descriptors in QSAR modeling based on bio-active conformations - PubMed [https://pubmed.ncbi.nlm.nih.gov/36617991/]
- 12. 3D-QSAR, design, molecular docking and dynamics ... [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2025.1545791/full]
- 13. 2D and 3D QSAR-Based Machine Learning Models for ... [https://www.sciencedirect.com/science/article/pii/S2468227625005381]
- 14. The three secrets to great 3D-QSAR: Alignment, alignment ... [https://cresset-group.com/about/news/the-three-secrets-to-great-3d-qsar-alignment-align/]
- 15. AutoGPA: An Automated 3D-QSAR Method Based on ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4207448/]
- 16. Py-CoMSIA: An Open-Source Implementation of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11945924/]
- 17. Automatic generation of alignments for 3D QSAR analyses [https://www.sciencedirect.com/science/article/abs/pii/S1093326301001103]
- 18. Field-based 3D-QSAR studies on amide- and urea ... [https://www.sciencedirect.com/science/article/abs/pii/S0169743923002344]
- 19. Automatic Generation of Alignments for 3D QSAR Analyses [https://pubmed.ncbi.nlm.nih.gov/11774998/]
- 20. Field based 3D QSAR (FQSAR) [https://bio-protocol.org/exchange/minidetail?id=907957&type=30]
- 21. Concepts and Applications Exemplified on Hydroxysteroid ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6332202/]
- 22. Pharmacophore modeling in drug design [https://www.sciencedirect.com/science/article/abs/pii/S1054358925000109]
- 23. Prioritization of new molecule design using QSAR models - 2D [https://cresset-group.com/discovery/specific/prioritization-new-molecule-design-qsar-models/]
- 24. Recent advances in molecular representation methods and ... [https://www.nature.com/articles/s44386-025-00017-2]
- 25. Knowledge-guided diffusion model for 3D ligand ... [https://www.nature.com/articles/s41467-025-57485-3]
- 26. Knowledge-guided diffusion model for 3D ligand ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11885826/]
- 27. 3D QSAR comparative molecular field analysis on ... [https://pubmed.ncbi.nlm.nih.gov/17055759/]
- 28. 3D-QSAR comparative molecular field analysis on opioid ... [https://pubmed.ncbi.nlm.nih.gov/15743203/]
- 29. Machine learning-based 3D-QSAR models for predicting ... [https://www.sciencedirect.com/science/article/pii/S3050787125000137]
- 30. Overview — Applications [https://docs.eyesopen.com/applications/rocs/rocs/rocs_overview.html]
- 31. FastROCS Shape based virtual screening software [https://www.eyesopen.com/fastrocs]
- 32. Shape-based screening - Schrödinger [https://www.schrodinger.com/life-science/learn/white-papers/shape-based-screening/]
- 33. ROCS - Shape Based Alignment for Virtual Screening [https://docs.eyesopen.com/floe/modules/small-molecule-modeling/docs/source/floes/ROCS_-_Shape_Based_Alignment_for_Virtual_Screening.html]
- 34. ChemBounce: a computational framework for scaffold hopping ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12449259/]
- 35. Derivation of Highly Predictive 3D-QSAR Models for hERG ... [https://www.mdpi.com/1424-8247/16/11/1509]
- 36. GRIND-based 3D-QSAR to predict inhibitory activity for ... [https://www.sciencedirect.com/science/article/abs/pii/S0223523407003583]
- 37. Alignment independent 3D-QSAR studies and molecular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7110456/]
- 38. Incorporating molecular shape into the alignment-free Grid- ... [https://documentsdelivered.com/source/012/100/012100727.php]
- 39. QSAR Accelerated Discovery of Potent Ice ... [https://www.nature.com/articles/srep26403]
- 40. Popular "Benchmark" Data Sets Do Not Distinguish the ... [https://pubmed.ncbi.nlm.nih.gov/19438212/]
- 41. 3D-QSAR, design, molecular docking and dynamics ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11841475/]
- 42. Development of novel monoamine oxidase B (MAO ... [https://pubmed.ncbi.nlm.nih.gov/35510607/]
- 43. Combining QSAR classification models for predictive ... [https://www.sciencedirect.com/science/article/abs/pii/S0223523412006393]
- 44. New Insights on the Activity and Selectivity of MAO-B ... [https://www.mdpi.com/1422-0067/24/11/9583]
- 45. Machine learning driven web-based app platform for the ... [https://www.nature.com/articles/s41598-024-55628-y]
- 46. Which conformations make stable crystal structures? Mapping ... [https://pubs.rsc.org/en/content/articlehtml/2014/sc/c4sc01132e]
- 47. Understanding and Quantifying Molecular Flexibility [https://pmc.ncbi.nlm.nih.gov/articles/PMC11523068/]
- 48. QSAR Modeling Based on Conformation Ensembles Using ... [https://pubmed.ncbi.nlm.nih.gov/34554736/]
- 49. A self-conformation-aware pre-training framework for ... [https://www.nature.com/articles/s41467-025-59634-0]
- 50. A precise comparison of molecular target prediction methods [https://pubs.rsc.org/en/content/articlehtml/2025/dd/d5dd00199d]
- 51. 3D-QSAR, Scaffold Hopping, Virtual Screening, and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11242256/]
- 52. Comparison of Different 2D and 3D-QSAR Methods on Activity ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3876572/]
- 53. Three-Dimensional-QSAR and Relative Binding Affinity ... [https://www.mdpi.com/1420-3049/28/3/1464]
- 54. Exploration of 2D and 3D-QSAR analysis and docking ... [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2023.1249041/full]
- 55. Py-CoMSIA: An Open-Source Implementation of ... [https://www.mdpi.com/1424-8247/18/3/440]
- 56. A Review of Machine Learning and QSAR/QSPR ... [https://www.mdpi.com/1420-3049/29/13/3159]
- 57. Consensus Modeling for HTS Assays Using In silico ... [https://www.frontiersin.org/journals/environmental-science/articles/10.3389/fenvs.2016.00002/full]
- 58. Py-CoMSIA: An Open-Source Implementation of ... [https://pubmed.ncbi.nlm.nih.gov/40143216/]
- 59. Comparison of various methods for validity evaluation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9396839/]
- 60. Beware of q2! [https://www.sciencedirect.com/science/article/abs/pii/S1093326301001231]
- 61. Robust and predictive 3D-QSAR models for ... - RSC Publishing [https://pubs.rsc.org/en/content/articlehtml/2024/ra/d4ra05342g]
- 62. Derivation of Highly Predictive 3D-QSAR Models for hERG ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10675541/]
- 63. Comparative study between deep learning and QSAR ... [https://www.nature.com/articles/s41598-020-73681-1]
- 64. Lead Optimization in Drug Discovery | Danaher Life Sciences [https://lifesciences.danaher.com/us/en/library/lead-optimization-drug-discovery-guide.html]
- 65. Lead Optimization in Drug Discovery [https://chem-space.com/news/chemspace-lead-otimization]
- 66. Efficient Drug Lead Discovery and Optimization - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC2727934/]
- 67. Navigating Lead Optimization in Drug Discovery - Cresset Group [https://cresset-group.com/postprocessing-scoring-metrics-2/]
- 68. Science Brief: 3D-QSAR Machine Learning for Binding ... [https://www.eyesopen.com/science-briefs/3d-qsar-machine-learning]
- 69. Lead Optimization [https://www.sartorius.com/en/applications/life-science-research/label-free-detection/lead-optimization]
- 70. Design, 3D-QSAR, molecular docking, MD simulations ... [https://www.sciencedirect.com/science/article/abs/pii/S0022286025000407]
- 71. 3D-QSAR Studies, Molecular Docking, Molecular Dynamic ... [https://www.mdpi.com/2075-1729/13/1/127]
- 72. Insights from 3D-QSAR, molecular docking, and dynamics for ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0326245]
- 73. atom-based 3D-QSAR, molecular docking, ADME-Tox, MD ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11827359/]
- 74. Unveiling Calotropis procera's promising wound healing ... [https://www.nature.com/articles/s41598-025-06822-z]
- 75. In silico design of novel potential isonicotinamide-based ... [https://pubs.rsc.org/en/content/articlelanding/2025/nj/d4nj01951b]